The promises and pitfalls of RNA-interference-based therapeutics
- PMID: 19158789
- PMCID: PMC2702667
- DOI: 10.1038/nature07758
The promises and pitfalls of RNA-interference-based therapeutics
Abstract
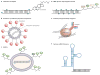
The discovery that gene expression can be controlled by the Watson-Crick base-pairing of small RNAs with messenger RNAs containing complementary sequence - a process known as RNA interference - has markedly advanced our understanding of eukaryotic gene regulation and function. The ability of short RNA sequences to modulate gene expression has provided a powerful tool with which to study gene function and is set to revolutionize the treatment of disease. Remarkably, despite being just one decade from its discovery, the phenomenon is already being used therapeutically in human clinical trials, and biotechnology companies that focus on RNA-interference-based therapeutics are already publicly traded.
Figures

References
-
- Zamore PD. RNA interference: big applause for silencing in Stockholm. Cell. 2006;127:1083–1086. - PubMed
-
-
McCaffrey AP, et al. RNA interference in adult mice. Nature. 2002;418:38–39.. This study was the first to show siRNA activity in vivo in mammals.
-
-
-
Song E, et al. RNA interference targeting Fas protects mice from fulminant hepatitis. Nature Med. 2003;9:347–351.. This paper provided the first therapeutic RNAi demonstration in animals.
-
-
-
Grimm D, et al. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541.. This article raised cautionary concerns about the danger of high-level shRNA expression in animals.
-
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

