Discovery of a non-peptidic inhibitor of west nile virus NS3 protease by high-throughput docking
- PMID: 19159012
- PMCID: PMC2613028
- DOI: 10.1371/journal.pntd.0000356
Discovery of a non-peptidic inhibitor of west nile virus NS3 protease by high-throughput docking
Abstract
Background: The non-structural 3 protease (NS3pro) is an essential flaviviral enzyme and therefore one of the most promising targets for drug development against West Nile virus (WNV) and dengue infections.
Methodology: In this work, a small-molecule inhibitor of the WNV NS3pro has been identified by automatic fragment-based docking of about 12000 compounds and testing by nuclear magnetic resonance (NMR) spectroscopy of only 22 molecules. Specific binding of the inhibitor into the active site of NS3pro and its binding mode are confirmed by 15N-HSQC NMR spectra. The inhibitory activity is further validated by an enzymatic assay and a tryptophan fluorescence quenching assay.
Conclusion: The inhibitor [4-(carbamimidoylsulfanylmethyl)-2,5-dimethylphenyl]-methylsulfanylmethanimidamide has a good ratio of binding affinity versus molecular weight (ligand efficiency of 0.33 kcal/mol per non-hydrogen atom), and thus has good potential as lead compound for further development to combat West Nile virus infections.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



 value derived from the fitting curves is about 40 µM (30±10 and 50±10 µM for the cross-peaks of Thr52 and Glu101, respectively).
value derived from the fitting curves is about 40 µM (30±10 and 50±10 µM for the cross-peaks of Thr52 and Glu101, respectively).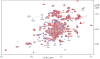
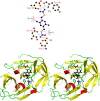
 helices in red,
helices in red,  strands in yellow, and loops in green. Atoms of compound 1 are colored as in the (top) with hydrogens in black. This figure was prepared using PyMOL (Delano Scientific, San Carlos, CA).
strands in yellow, and loops in green. Atoms of compound 1 are colored as in the (top) with hydrogens in black. This figure was prepared using PyMOL (Delano Scientific, San Carlos, CA).
References
-
- Division of Vector-Borne Infectious Disease. West Nile virus homepage. 2007. Centers for Disease Control and Prevention. http://www.cdc.gov/ncidod/dvbid/westnile.
-
- Hayes EB, Gubler DJ. West Nile virus: epidemiology and clinical features of an emerging epidemic in the United States. Ann Rev Med. 2006;57:181–194. - PubMed
-
- Mukhopadhyay S, Kuhn RJ, Rossmann MG. A structural perspective of the flavivirus life cycle. Nat Rev Microbiol. 2005;3:13–22. - PubMed
-
- Chappell KJ, Stoermer MJ, Fairlie DP, Young PR. Insights to substrate binding and processing by West Nile Virus NS3 protease through combined modeling, protease mutagenesis, and kinetic studies. J Biol Chem. 2006;281:38448–38458. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

