New DArT markers for oat provide enhanced map coverage and global germplasm characterization
- PMID: 19159465
- PMCID: PMC2661094
- DOI: 10.1186/1471-2164-10-39
New DArT markers for oat provide enhanced map coverage and global germplasm characterization
Abstract
Background: Genomic discovery in oat and its application to oat improvement have been hindered by a lack of genetic markers common to different genetic maps, and by the difficulty of conducting whole-genome analysis using high-throughput markers. This study was intended to develop, characterize, and apply a large set of oat genetic markers based on Diversity Array Technology (DArT).
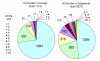
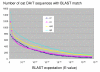
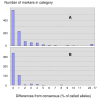
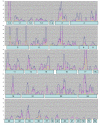
Results: Approximately 19,000 genomic clones were isolated from complexity-reduced genomic representations of pooled DNA samples from 60 oat varieties of global origin. These were screened on three discovery arrays, with more than 2000 polymorphic markers being identified for use in this study, and approximately 2700 potentially polymorphic markers being identified for use in future studies. DNA sequence was obtained for 2573 clones and assembled into a non-redundant set of 1770 contigs and singletons. Of these, 705 showed highly significant (Expectation < 10E-10) BLAST similarity to gene sequences in public databases. Based on marker scores in 80 recombinant inbred lines, 1010 new DArT markers were used to saturate and improve the 'Kanota' x 'Ogle' genetic map. DArT markers provided map coverage approximately equivalent to existing markers. After binning markers from similar clones, as well as those with 99% scoring similarity, a set of 1295 non-redundant markers was used to analyze genetic diversity in 182 accessions of cultivated oat of worldwide origin. Results of this analysis confirmed that major clusters of oat diversity are related to spring vs. winter type, and to the presence of major breeding programs within geographical regions. Secondary clusters revealed groups that were often related to known pedigree structure.
Conclusion: These markers will provide a solid basis for future efforts in genomic discovery, comparative mapping, and the generation of an oat consensus map. They will also provide new opportunities for directed breeding of superior oat varieties, and guidance in the maintenance of oat genetic diversity.
Figures



References
-
- Baum BR. Oats: wild and cultivated. A monograph of the genus Avena L. (Poaceae). Monograph No. 14. Ottawa, Canada: Biosystematics Research Institute (curently ECORC), Agriculture and Agri-Food Canada; 1977.
-
- Leggett JM. Classification and speciation in Avena. In: Sorrells ME, Marshall HG, editor. Oat Science and Technology, Agronomy Monograph 33. Madison, WI: American Society of Agronomy and Crop Science Society of America; 1992. pp. 29–52.
-
- Chao S, Zhang W, Dubcovsky J, Sorrells M. Evaluation of genetic diversity and genome-wide linkage disequilibrium among US wheat (Triticum aestivum L.) germplasm representing different market classes. Crop Sci. 2007;47:1018–1030.
-
- Fu YB, Peterson GW, Williams D, Richards KW, Fetch JM. Patterns of AFLP variation in a core subset of cultivated hexaploid oat germplasm. Theor Appl Genet. 2005;111:530–539. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

