Molecular models of human P-glycoprotein in two different catalytic states
- PMID: 19159494
- PMCID: PMC2661087
- DOI: 10.1186/1472-6807-9-3
Molecular models of human P-glycoprotein in two different catalytic states
Abstract
Background: P-glycoprotein belongs to the family of ATP-binding cassette proteins which hydrolyze ATP to catalyse the translocation of their substrates through membranes. This protein extrudes a large range of components out of cells, especially therapeutic agents causing a phenomenon known as multidrug resistance. Because of its clinical interest, its activity and transport function have been largely characterized by various biochemical studies. In the absence of a high-resolution structure of P-glycoprotein, homology modeling is a useful tool to help interpretation of experimental data and potentially guide experimental studies.
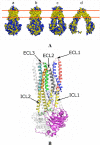
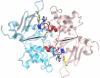
Results: We present here three-dimensional models of two different catalytic states of P-glycoprotein that were developed based on the crystal structures of two bacterial multidrug transporters. Our models are supported by a large body of biochemical data. Measured inter-residue distances correlate well with distances derived from cross-linking data. The nucleotide-free model features a large cavity detected in the protein core into which ligands of different size were successfully docked. The locations of docked ligands compare favorably with those suggested by drug binding site mutants.
Conclusion: Our models can interpret the effects of several mutants in the nucleotide-binding domains (NBDs), within the transmembrane domains (TMDs) or at the NBD:TMD interface. The docking results suggest that the protein has multiple binding sites in agreement with experimental evidence. The nucleotide-bound models are exploited to propose different pathways of signal transmission upon ATP binding/hydrolysis which could lead to the elaboration of conformational changes needed for substrate translocation. We identified a cluster of aromatic residues located at the interface between the NBD and the TMD in opposite halves of the molecule which may contribute to this signal transmission. Our models may characterize different steps in the catalytic cycle and may be important tools to understand the structure-function relationship of P-glycoprotein.
Figures



References
-
- Dean M, Hamon Y, Chimini G. The human ATP-binding cassette (ABC) transporter superfamily. J Lipid Res. 2001;42:1007–1017. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

