Toward accurate screening in computer-aided enzyme design
- PMID: 19161327
- PMCID: PMC2832760
- DOI: 10.1021/bi802191b
Toward accurate screening in computer-aided enzyme design
Abstract
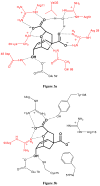
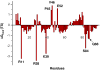
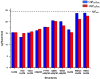
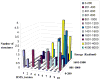
The ability to design effective enzymes is one of the most fundamental challenges in biotechnology and in some respects in biochemistry. In fact, such ability would be one of the most convincing manifestations of a full understanding of the origin of enzyme catalysis. In this work, we explore the reliability of different simulation approaches, in terms of their ability to rank different possible active site constructs. This validation is done by comparing the ability of different approaches to evaluate the catalytic contributions of various residues in chorismate mutase. It is demonstrated that the empirical valence bond (EVB) model can serve as a practical yet accurate tool in the final stages of computer-aided enzyme design (CAED). Other approaches for fast screening are also examined and found to be less accurate and mainly useful for qualitative screening of ionized residues. It is pointed out that accurate ranking of different options for enzyme design cannot be accomplished by approaches that cannot capture the electrostatic preorganization effect. This is in particular true with regard to current design approaches that use gas phase or small cluster calculations and then estimate the interaction between the enzyme and the transition state (TS) model rather than the TS binding free energy or the relevant activation free energy. The ability of the EVB model to provide a tool for quantitative ranking in the final stage of CAED may help in progressing toward the design of enzymes whose catalytic power is closer to that of native enzymes than to that of the current generation of designer enzymes.
Figures
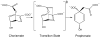
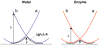
References
-
- Toscano MD, Woycechowsky KJ, Hilvert D. Minimalist active-site redesign: Teaching old enzymes new tricks. Angew Chem Int Ed. 2007;46:4468–4470. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

