What are microarrays teaching us about sleep?
- PMID: 19162550
- PMCID: PMC2942088
- DOI: 10.1016/j.molmed.2008.12.002
What are microarrays teaching us about sleep?
Abstract
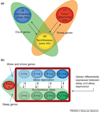
Many fundamental questions about sleep remain unanswered. The presence of sleep across phyla suggests that it must serve a basic cellular and/or molecular function. Microarray studies, performed in several model systems, have identified classes of genes that are sleep-state regulated. This has led to the following concepts: first, a function of sleep is to maintain synaptic homeostasis; second, sleep is a stage of macromolecule biosynthesis; third, extending wakefulness leads to downregulation of several important metabolic pathways; and, fourth, extending wakefulness leads to endoplasmic reticulum stress. In human studies, microarrays are being applied to the identification of biomarkers for sleepiness and for the common debilitating condition of obstructive sleep apnea.
Figures

References
-
- Mackiewicz M, Pack AI. Functional genomics of sleep. Respir. Physiol. Neurobiol. 2003;135:207–220. - PubMed
-
- Rhyner TA, et al. Molecular cloning of forebrain mRNAs which are modulated by sleep deprivation. Eur. J. Neurosci. 1990;2:1063–1073. - PubMed
-
- Liang P, Pardee AB. Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science. 1992;257:967–971. - PubMed
-
- Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

