Whole-genome approach implicates CD44 in cellular resistance to carboplatin
- PMID: 19164090
- PMCID: PMC2683878
- DOI: 10.1186/1479-7364-3-2-128
Whole-genome approach implicates CD44 in cellular resistance to carboplatin
Abstract
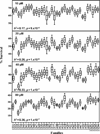
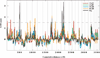
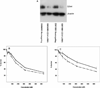
Carboplatin is a chemotherapeutic agent used in the management of many cancers, yet treatment is limited by resistance and toxicities. To achieve a better understanding of the genetic contribution to carboplatin resistance or toxicities, lymphoblastoid cell lines from 34 large Centre d'Etude du Polymorphisme Humain pedigrees were utilised to evaluate interindividual variation in carboplatin cytotoxicity. Significant heritability, ranging from 0.17-0.36 (p = 1 x 10(-7) to 9 x 10(-4)), was found for cell growth inhibition following 72-hour treatment at each carboplatin concentration (10, 20, 40 and 80 microM) and IC(50) (concentration for 50 per cent cell growth inhibition). Linkage analysis revealed 11 regions with logarithm of odds (LOD) scores greater than 1.5. The highest LOD score on chromosome 11 (LOD = 3.36, p = 4.2 x 10(-5)) encompasses 65 genes within the 1 LOD confidence interval for the carboplatin IC 50 . We further analysed the IC(50) phenotype with a linkage-directed association analysis using 71 unrelated HapMap and Perlegen cell lines and identified 18 single nucleotide polymorphisms within eight genes that were significantly associated with the carboplatin IC(50) (p < 3.6 x 10(-5); false discovery rate <5 per cent). Next, we performed linear regression on the baseline expression and carboplatin IC(50) values of the eight associated genes, which identified the most significant correlation between CD44 expression and IC(50) (r(2)= 0.20; p = 6 x 10(-4)). The quantitative real-time polymerase chain reaction further confirmed a statistically significant difference in CD44 expression levels between carboplatin-resistant and -sensitive cell lines (p = 5.9 x 10(-3)). Knockdown of CD44 expression through small interfering RNA resulted in increased cellular sensitivity to carboplatin (p < 0.01). Our whole-genome approach using molecular experiments identified CD44 as being important in conferring cellular resistance to carboplatin.
Figures


References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

