Genetic analysis of factors affecting susceptibility of Bacillus subtilis to daptomycin
- PMID: 19164152
- PMCID: PMC2663116
- DOI: 10.1128/AAC.01329-08
Genetic analysis of factors affecting susceptibility of Bacillus subtilis to daptomycin
Abstract
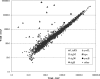
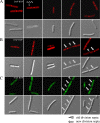
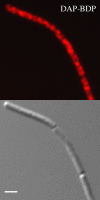
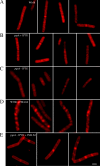
Daptomycin is the first of a new class of cyclic lipopeptide antibiotics used against multidrug-resistant, gram-positive pathogens. The proposed mechanism of action involves disruption of the functional integrity of the bacterial membrane in a Ca(2+)-dependent manner. We have used transcriptional profiling to demonstrate that treatment of Bacillus subtilis with daptomycin strongly induces the lia operon including the autoregulatory LiaRS two-component system (homologous to Staphylococcus aureus VraSR). The lia operon protects against daptomycin, and deletion of liaH, encoding a phage-shock protein A (PspA)-like protein, leads to threefold increased susceptibility. Since daptomycin interacts with the membrane, we tested mutants with altered membrane composition for effects on susceptibility. Deletion mutations of mprF (lacking lysyl-phosphatidylglycerol) or des (lipid desaturase) increased daptomycin susceptibility, whereas overexpression of MprF decreased susceptibility. Conversely, depletion of the cell for the anionic lipid phosphatidylglycerol led to increased resistance. Fluorescently labeled daptomycin localized to the septa and in a helical pattern around the cell envelope and was delocalized upon the depletion of phosphatidylglycerol. Together, these results indicate that the daptomycin-Ca(2+) complex interacts preferentially with regions enriched in anionic phospholipids and leads to membrane stresses that can be ameliorated by PspA family proteins.
Figures

References
-
- Antelmann, H., S. Towe, D. Albrecht, and M. Hecker. 2007. The phosphorus source phytate changes the composition of the cell wall proteome in Bacillus subtilis. J. Proteome Res. 6:897-903. - PubMed
-
- Bisicchia, P., D. Noone, E. Lioliou, A. Howell, S. Quigley, T. Jensen, H. Jarmer, and K. M. Devine. 2007. The essential YycFG two-component system controls cell wall metabolism in Bacillus subtilis. Mol. Microbiol. 65:180-200. - PubMed
-
- Butcher, B. G., and J. D. Helmann. 2006. Identification of Bacillus subtilis sigma-dependent genes that provide intrinsic resistance to antimicrobial compounds produced by bacilli. Mol. Microbiol. 60:765-782. - PubMed
-
- Campo, N., H. Tjalsma, G. Buist, D. Stepniak, M. Meijer, M. Veenhuis, M. Westermann, J. P. Muller, S. Bron, J. Kok, O. P. Kuipers, and J. D. Jongbloed. 2004. Subcellular sites for bacterial protein export. Mol. Microbiol. 53:1583-1599. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

