Grade-dependent proteomics characterization of kidney cancer
- PMID: 19164279
- PMCID: PMC2689781
- DOI: 10.1074/mcp.M800252-MCP200
Grade-dependent proteomics characterization of kidney cancer
Abstract
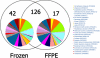
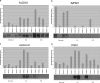
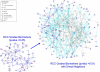
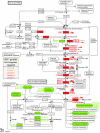
Kidney cancer is frequently metastatic on presentation at which point the disease is associated with a 95% mortality. Assessment of tumor grade on pathological examination is the most powerful means for prognostication as well as for stratification of patients into those who might respond to conventional or targeted therapy. Although there exist several grading systems in common use, all suffer from significant disparity among observers. In an attempt to objectify this process as well as to acquire grade-specific mechanistic information, we performed LC-MS/MS-based proteomics analysis on 50 clear cell kidney cancers equally distributed among normal tissues and Fuhrman grades 1-4. Initial experiments confirmed the utility of using archived formalin-fixed paraffin-embedded samples for LC-MS/MS-based proteomics analysis, and the LC-MS/MS findings were validated by extensive immunoblotting. We now show that changes among many biochemical processes and pathways are strongly grade-dependent with the glycolytic and amino acid synthetic pathways highly represented. In addition, proteins relating to acute phase and xenobiotic metabolism signaling are highly represented. Self-organized mapping of proteins with similar patterns of expression led to the creation of a heat map that will be useful in grade characterization as well as in future research relating to oncogenic mechanisms and targeted therapies for kidney cancer.
Figures




References
-
- Weiss, R. H., and Lin, P.-Y. ( 2006) Kidney cancer: identification of novel targets for therapy. Kidney Int. 69, 224–232 - PubMed
-
- Novara, G., Martignoni, G., Artibani, W., and Ficarra, V. ( 2007) Grading systems in renal cell carcinoma. J. Urol. 177, 430–436 - PubMed
-
- Fuhrman, S. A., Lasky, L. C., and Limas, C. ( 1982) Prognostic significance of morphologic parameters in renal cell carcinoma. Am. J. Surg. Pathol. 6, 655–663 - PubMed
-
- Hwang, S. I., Thumar, J., Lundgren, D. H., Rezaul, K., Mayya, V., Wu, L., Eng, J., Wright, M. E., and Han, D. K. ( 2007) Direct cancer tissue proteomics: a method to identify candidate cancer biomarkers from formalin-fixed paraffin-embedded archival tissues. Oncogene 26, 65–76 - PubMed
-
- Wessel, D., and Flugge, U. I. ( 1984) A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 138, 141–143 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

