Chloroplast protein targeting involves localized translation in Chlamydomonas
- PMID: 19164529
- PMCID: PMC2629442
- DOI: 10.1073/pnas.0811268106
Chloroplast protein targeting involves localized translation in Chlamydomonas
Abstract
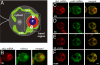
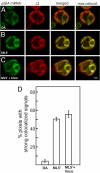
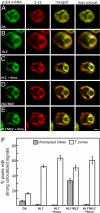
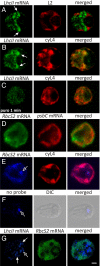
The compartmentalization of eukaryotic cells requires that newly synthesized proteins be targeted to the compartments in which they function. In chloroplasts, a few thousand proteins function in photosynthesis, expression of the chloroplast genome, and other processes. Most chloroplast proteins are synthesized in the cytoplasm, imported, and then targeted to a specific chloroplast compartment. The remainder are encoded by the chloroplast genome, synthesized within the organelle, and targeted by mechanisms that are only beginning to be elucidated. We used fluorescence confocal microscopy to explore the targeting mechanisms used by several chloroplast proteins in the green alga Chlamydomonas. These include the small subunit of ribulose bisphosphate carboxylase (rubisco) and the light-harvesting complex II (LHCII) subunits, which are imported from the cytoplasm, and 2 proteins synthesized in the chloroplast: the D1 subunit of photosystem II and the rubisco large subunit. We determined whether the targeting of each protein involves localized translation of the mRNA that encodes it. When this was the case, we explored whether the targeting sequence was in the nascent polypeptide or in the mRNA, based on whether the localization was translation-dependent or -independent, respectively. The results reveal 2 novel examples of targeting by localized translation, in LHCII subunit import and the targeting of the rubisco large subunit to the pyrenoid. They also demonstrate examples of each of the three known mechanisms-posttranslational, cotranslational (signal recognition particle-mediated), and mRNA-based-in the targeting of specific chloroplast proteins. Our findings can help guide the exploration of these pathways at the biochemical level.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Photosystem II assembly and repair are differentially localized in Chlamydomonas.Plant Cell. 2007 Nov;19(11):3640-54. doi: 10.1105/tpc.107.054882. Epub 2007 Nov 30. Plant Cell. 2007. PMID: 18055604 Free PMC article.
-
Photosystem Biogenesis Is Localized to the Translation Zone in the Chloroplast of Chlamydomonas.Plant Cell. 2019 Dec;31(12):3057-3072. doi: 10.1105/tpc.19.00263. Epub 2019 Oct 7. Plant Cell. 2019. PMID: 31591163 Free PMC article.
-
Chloroplast biogenesis involves spatial coordination of nuclear and organellar gene expression in Chlamydomonas.Plant Physiol. 2024 Sep 2;196(1):112-123. doi: 10.1093/plphys/kiae256. Plant Physiol. 2024. PMID: 38709497 Free PMC article.
-
Senescence-associated degradation of chloroplast proteins inside and outside the organelle.Plant Biol (Stuttg). 2008 Sep;10 Suppl 1:15-22. doi: 10.1111/j.1438-8677.2008.00089.x. Plant Biol (Stuttg). 2008. PMID: 18721308 Review.
-
Synthesis, membrane insertion and assembly of the chloroplast-encoded D1 protein into photosystem II.FEBS Lett. 2002 Feb 13;512(1-3):13-8. doi: 10.1016/s0014-5793(02)02218-4. FEBS Lett. 2002. PMID: 11852043 Review.
Cited by
-
Studies on subcellular compartmentalization of plant pathogenic noncoding RNAs give new insights into the intracellular RNA-traffic mechanisms.Plant Physiol. 2012 Jun;159(2):558-64. doi: 10.1104/pp.112.195214. Epub 2012 Apr 3. Plant Physiol. 2012. PMID: 22474218 Free PMC article. No abstract available.
-
Native architecture of the Chlamydomonas chloroplast revealed by in situ cryo-electron tomography.Elife. 2015 Jan 13;4:e04889. doi: 10.7554/eLife.04889. Elife. 2015. PMID: 25584625 Free PMC article.
-
Evolutionary origins of metabolic compartmentalization in eukaryotes.Philos Trans R Soc Lond B Biol Sci. 2010 Mar 12;365(1541):847-55. doi: 10.1098/rstb.2009.0252. Philos Trans R Soc Lond B Biol Sci. 2010. PMID: 20124349 Free PMC article. Review.
-
New insights in the topology of the biosynthesis of 5-aminolevulinic acid.Plant Signal Behav. 2013 Feb;8(2):e23124. doi: 10.4161/psb.23124. Epub 2013 Jan 8. Plant Signal Behav. 2013. PMID: 23299429 Free PMC article. Review.
-
A prion-like domain is required for phase separation and chloroplast RNA processing during cold acclimation in Arabidopsis.Plant Cell. 2024 Jul 31;36(8):2851-2872. doi: 10.1093/plcell/koae145. Plant Cell. 2024. PMID: 38723165 Free PMC article.
References
-
- St. Johnston D. Moving messages: The intracellular localization of mRNAs. Nat Rev Mol Cell Biol. 2005;6:363–375. - PubMed
-
- Stephens SB, et al. Analysis of mRNA partitioning between the cytosol and endoplasmic reticulum compartments of mammalian cells. Methods Mol Biol. 2008;419:197–214. - PubMed
-
- Inaba T, Schnell DJ. Protein trafficking to plastids: One theme, many variations. Biochem J. 2008;413:15–28. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

