Effects of linker sequences on vesicle fusion mediated by lipid-anchored DNA oligonucleotides
- PMID: 19164559
- PMCID: PMC2633564
- DOI: 10.1073/pnas.0812356106
Effects of linker sequences on vesicle fusion mediated by lipid-anchored DNA oligonucleotides
Abstract
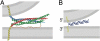
Synthetic lipid-oligonucleotide conjugates inserted into lipid vesicles mediate fusion when one population of vesicles displays the 5'-coupled conjugate and the other the 3'-coupled conjugate, so that anti-parallel hybridization allows the membrane surfaces to come into close proximity. Improved assays show that lipid mixing proceeds more quickly and to a much greater extent than content mixing, suggesting the latter is rate limiting. To test the effect of membrane-membrane spacing on fusion, a series of conjugates was constructed by adding 2-24 noncomplementary bases at the membrane-proximal ends of two complementary sequences. Increasing linker lengths generally resulted in progressively reduced rates and extents of lipid and content mixing, in contrast to higher vesicle docking rates. The relatively flexible, single-stranded DNA linker facilitates docking but allows greater spacing between the vesicles after docking, thus making the transition into fusion less probable, but not preventing it altogether. These experiments demonstrate the utility of DNA as a model system for fusion proteins, where sequence can easily be modified to systematically probe the effect of distance between bilayers in the fusion reaction.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
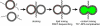


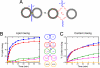

References
-
- Chen YA, Scheller RH. SNARE-mediated membrane fusion. Nat Rev Mol Cell Biol. 2001;2:98–106. - PubMed
-
- Jahn R, Lang T, Südhof TC. Membrane fusion. Cell. 2003;112:519–533. - PubMed
-
- Brunger AT. Structure and function of SNARE and SNARE-interacting proteins. Q Rev Biophys. 2005;38:1–47. - PubMed
-
- Weber T, et al. SNAREpins: Minimal machinery for membrane fusion. Cell. 1998;92:759–772. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

