RodZ, a component of the bacterial core morphogenic apparatus
- PMID: 19164570
- PMCID: PMC2633561
- DOI: 10.1073/pnas.0810794106
RodZ, a component of the bacterial core morphogenic apparatus
Abstract
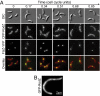
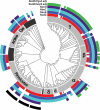
The molecular basis of bacterial cell morphogenesis remains largely an open question. Here we discover a morphogenic protein, RodZ, which is widely conserved across the bacterial kingdom. In Caulobacter crescentus, RodZ is essential for viability and is involved in all aspects of this organism's complex morphology. Depletion or over-production of RodZ results in grossly misshapen cells with stalk defects. RodZ exhibits a localization pattern during the cell cycle corresponding to sites of active peptidoglycan synthesis. The temporal transition of RodZ between patchy/helical and mid-cell localization mimics and depends on the actin-like MreB cytoskeleton. In Escherichia coli, an organism with a distinct mode of growth and MreB localization dynamics, RodZ follows MreB and retains its crucial role in cell morphogenesis, demonstrating conservation of function. Genomic analysis shows that RodZ represents an ancient function unique to bacteria. Multiple sequence alignment of 143 RodZ sequences from species across bacterial phyla identifies an N-terminal cytoplasmic domain with a helix-turn-helix motif, a transmembrane sequence, and a previously unidentified, conserved periplasmic or extracellular C-terminal domain. Both the N- and C-terminal domains are important for function, with the N-terminal domain containing localization determinants. This study uncovers a key missing player in the cytoskeleton-based growth machinery enabling heritable and defined cellular forms in bacteria.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

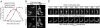


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

