Intersubunit allosteric communication mediated by a conserved loop in the MCM helicase
- PMID: 19164574
- PMCID: PMC2633543
- DOI: 10.1073/pnas.0809192106
Intersubunit allosteric communication mediated by a conserved loop in the MCM helicase
Abstract
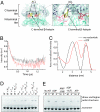
The minichromosome maintenance (MCM) helicase is the presumptive replicative helicase in archaea and eukaryotes. The archaeal homomultimeric MCM has a two-tier structure. One tier contains the AAA+ motor domains of the proteins, and these are the minimal functional helicase domains. The second tier is formed by the N-terminal domains. These domains are not essential for MCM helicase activity but act to enhance the processivity of the helicase. We reveal that a conserved loop facilitates communication between processivity and motor tiers. Interestingly, this allostery seems to be mediated by interactions between, rather than within, individual protomers in the MCM ring.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


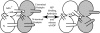
References
-
- Costa A, Onesti S. The MCM complex: (Just) a replicative helicase? Biochem Soc Trans. 2008;36:136–140. - PubMed
-
- Fletcher RJ, et al. The structure and function of MCM from archaeal M. thermoautotrophicum. Nat Struct Biol. 2003;10:160–167. - PubMed
-
- McGeoch AT, Trakselis MA, Laskey RA, Bell SD. Organization of the archaeal MCM complex on DNA and implications for the helicase mechanism. Nat Struct Mol Biol. 2005;12:756–762. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

