Polymicrobial interactions stimulate resistance to host innate immunity through metabolite perception
- PMID: 19164580
- PMCID: PMC2629492
- DOI: 10.1073/pnas.0809533106
Polymicrobial interactions stimulate resistance to host innate immunity through metabolite perception
Abstract
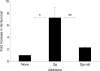
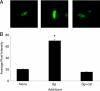
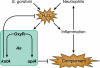
Bacteria in the human oral cavity often grow in an attached multispecies biofilm community. Members of this community display defined interactions that have an impact on the physiology of the individual and the group. Here, we show that during coculture growth with streptococci, the oral pathogen Aggregatibacter actinomycetemcomitans displays enhanced resistance to killing by host innate immunity. The mechanism of resistance involves sensing of the streptococcal metabolite hydrogen peroxide by A. actinomycetemcomitans, which stimulates a genetic program resulting in enhanced expression of the complement resistance protein ApiA. The oxidative stress response regulator OxyR mediates induction of apiA transcription, and this induction is required for coculture resistance to killing by human serum. These findings provide evidence that interaction between community members mediates prokaryotic resistance to host innate immunity and reinforce the need to understand how polymicrobial growth affects interaction with the host immune system.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Meyer DH, Fives-Taylor PM. Oral pathogens: from dental plaque to cardiac disease. Curr Opin Microbiol. 1998;1:88–95. - PubMed
-
- Ebersole JL, Cappelli D, Sandoval MN. Subgingival distribution of A. actinomycetemcomitans in periodontitis. J Clin Periodontol. 1994;21:65–75. - PubMed
-
- Courts FJ, Boackle RJ, Fudenberg HH, Silverman MS. Detection of functional complement components in gingival crevicular fluid from humans with periodontal diseases. J Dent Res. 1977;56:327–331. - PubMed
-
- Biswas S, Duperon DF, Chebib FS. Study of crevice fluid in relation to periodontal disease in children. II. Effect of age, sex and gingival inflammation on crevice fluid protein, carbohydrate, total calcium, phosphate and nitrogen. J Periodontal Res. 1977;12:265–278. - PubMed
-
- Ficara AJ, Levin MP, Grower MF, Kramer GD. A comparison of the glucose and protein content of gingival fluid from diabetics and nondiabetics. J Periodontal Res. 1975;10:171–175. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

