A structural overview of the vertebrate prion proteins
- PMID: 19164911
- PMCID: PMC2634592
- DOI: 10.4161/pri.1.3.5281
A structural overview of the vertebrate prion proteins
Abstract
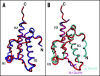
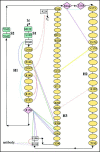
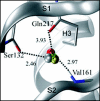
Among the diseases caused by protein misfolding is the family associated with the prion protein (PrP). This is a small extracellular membrane-anchored molecule of yet unknown function. Understanding how PrP folds both into its cellular and pathological forms is thought to be crucial for explaining protein misfolding in general and the specific role of PrP in disease. Since the first structure determination, an increasing number of structural studies of PrP have become available, showing that the protein is formed by a flexible N-terminal region and a highly conserved globular C-terminal domain. We review here the current knowledge on PrP structure. We focus on vertebrate PrPs and analyse in detail the similarities and the differences among the coordinates of the C-terminal domain of PrP from different species, in search for understanding the mechanism of disease-causing mutations and the molecular bases of species barrier.
Figures






Similar articles
-
Selective processing and metabolism of disease-causing mutant prion proteins.PLoS Pathog. 2009 Jun;5(6):e1000479. doi: 10.1371/journal.ppat.1000479. Epub 2009 Jun 19. PLoS Pathog. 2009. PMID: 19543376 Free PMC article.
-
Impact of methionine oxidation as an initial event on the pathway of human prion protein conversion.Prion. 2013 Sep-Oct;7(5):404-11. doi: 10.4161/pri.26745. Epub 2013 Oct 9. Prion. 2013. PMID: 24121542 Free PMC article.
-
Pathogenic mutations within the hydrophobic domain of the prion protein lead to the formation of protease-sensitive prion species with increased lethality.J Virol. 2014 Mar;88(5):2690-703. doi: 10.1128/JVI.02720-13. Epub 2013 Dec 18. J Virol. 2014. PMID: 24352465 Free PMC article.
-
Structural factors underlying the species barrier and susceptibility to infection in prion disease.Biochem Cell Biol. 2010 Apr;88(2):195-202. doi: 10.1139/o09-172. Biochem Cell Biol. 2010. PMID: 20453922 Review.
-
Prion neurotoxicity: insights from prion protein mutants.Curr Issues Mol Biol. 2010;12(2):51-61. Epub 2009 Sep 18. Curr Issues Mol Biol. 2010. PMID: 19767650 Free PMC article. Review.
Cited by
-
Human Prion Disorders: Review of the Current Literature and a Twenty-Year Experience of the National Surveillance Center in the Czech Republic.Diagnostics (Basel). 2021 Oct 1;11(10):1821. doi: 10.3390/diagnostics11101821. Diagnostics (Basel). 2021. PMID: 34679519 Free PMC article. Review.
-
Prion peptide uptake in microglial cells--the effect of naturally occurring autoantibodies against prion protein.PLoS One. 2013 Jun 28;8(6):e67743. doi: 10.1371/journal.pone.0067743. Print 2013. PLoS One. 2013. PMID: 23840767 Free PMC article.
-
Prion protein "gamma-cleavage": characterizing a novel endoproteolytic processing event.Cell Mol Life Sci. 2016 Feb;73(3):667-83. doi: 10.1007/s00018-015-2022-z. Epub 2015 Aug 23. Cell Mol Life Sci. 2016. PMID: 26298290 Free PMC article.
-
Membrane Domain Localization and Interaction of the Prion-Family Proteins, Prion and Shadoo with Calnexin.Membranes (Basel). 2021 Dec 13;11(12):978. doi: 10.3390/membranes11120978. Membranes (Basel). 2021. PMID: 34940479 Free PMC article.
-
Do prion protein gene polymorphisms induce apoptosis in non-mammals?J Biosci. 2016 Mar;41(1):97-107. doi: 10.1007/s12038-015-9584-7. J Biosci. 2016. PMID: 26949092
References
-
- Prusiner SB. Novel proteinaceous infectious particles cause scrapie. Science. 1982;216:136–144. - PubMed
-
- Prusiner SB. Prions are novel infectious pathogens causing scrapie and Creutzfeldt-Jakob disease. Bioessays. 1986;5:281–286. - PubMed
-
- Prusiner SB. Human prion diseases and neurodegeneration. Curr Top Microbiol Immunol. 1996;207:1–17. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
