Retroviral integrase superfamily: the structural perspective
- PMID: 19165139
- PMCID: PMC2637324
- DOI: 10.1038/embor.2008.256
Retroviral integrase superfamily: the structural perspective
Abstract
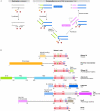
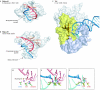
The retroviral integrase superfamily (RISF) comprises numerous important nucleic acid-processing enzymes, including transposases, integrases and various nucleases. These enzymes are involved in a wide range of processes such as transposition, replication and repair of DNA, homologous recombination, and RNA-mediated gene silencing. Two out of the four enzymes that are encoded by the human immunodeficiency virus--RNase H1 and integrase--are members of this superfamily. RISF enzymes act on various substrates, and yet show remarkable mechanistic and structural similarities. All share a common fold of the catalytic core and the active site, which is composed primarily of carboxylate residues. Here, I present RISF proteins from a structural perspective, describing the individual members and the common and divergent elements of their structures, as well as the mechanistic insights gained from the structures of RNase H1 enzyme complexes with RNA/DNA hybrids.
Figures

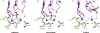

References
-
- Ariyoshi M, Vassylyev DG, Iwasaki H, Nakamura H, Shinagawa H, Morikawa K (1994) Atomic structure of the RuvC resolvase: a Holliday junction-specific endonuclease from E. coli. Cell 78: 1063–1072 - PubMed
-
- Bennett RJ, Dunderdale HJ, West SC (1993) Resolution of Holliday junctions by RuvC resolvase: cleavage specificity and DNA distortion. Cell 74: 1021–1031 - PubMed
-
- Cerritelli SM, Frolova EG, Feng C, Grinberg A, Love PE, Crouch RJ (2003) Failure to produce mitochondrial DNA results in embryonic lethality in Rnaseh1 null mice. Mol Cell 11: 807–815 - PubMed
-
- Chapados BR, Chai Q, Hosfield DJ, Qiu J, Shen B, Tainer JA (2001) Structural biochemistry of a type 2 RNase H: RNA primer recognition and removal during DNA replication. J Mol Biol 307: 541–556 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

