dsRNA with 5' overhangs contributes to endogenous and antiviral RNA silencing pathways in plants
- PMID: 19165150
- PMCID: PMC2657584
- DOI: 10.1038/emboj.2009.2
dsRNA with 5' overhangs contributes to endogenous and antiviral RNA silencing pathways in plants
Abstract
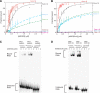
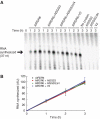
In plants, SGS3 and RNA-dependent RNA polymerase 6 (RDR6) are required to convert single- to double-stranded RNA (dsRNA) in the innate RNAi-based antiviral response and to produce both exogenous and endogenous short-interfering RNAs. Although a role for RDR6-catalysed RNA-dependent RNA polymerisation in these processes seems clear, the function of SGS3 is unknown. Here, we show that SGS3 is a dsRNA-binding protein with unexpected substrate selectivity favouring 5'-overhang-containing dsRNA. The conserved XS and coiled-coil domains are responsible for RNA-binding activity. Furthermore, we find that the V2 protein from tomato yellow leaf curl virus, which suppresses the RNAi-based host immune response, is a dsRNA-binding protein with similar specificity to SGS3. In competition-binding experiments, V2 outcompetes SGS3 for substrate dsRNA recognition, whereas a V2 point mutant lacking the suppressor function in vivo cannot efficiently overcome SGS3 binding. These findings suggest that SGS3 recognition of dsRNA containing a 5' overhang is required for subsequent steps in RNA-mediated gene silencing in plants, and that V2 functions as a viral suppressor by preventing SGS3 from accessing substrate RNAs.
Figures





References
-
- Allen E, Xie Z, Gustafson AM, Carrington JC (2005) microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121: 207–221 - PubMed
-
- Beclin C, Boutet S, Waterhouse P, Vaucheret H (2002) A branched pathway for transgene-induced RNA silencing in plants. Curr Biol 12: 684–688 - PubMed
-
- Burkhard P, Stetefeld J, Strelkov SV (2001) Coiled coils: a highly versatile protein folding motif. Trends Cell Biol 11: 82–88 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

