Inflammatory genital infections mitigate a severe genetic bottleneck in heterosexual transmission of subtype A and C HIV-1
- PMID: 19165325
- PMCID: PMC2621345
- DOI: 10.1371/journal.ppat.1000274
Inflammatory genital infections mitigate a severe genetic bottleneck in heterosexual transmission of subtype A and C HIV-1
Abstract
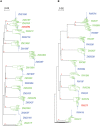
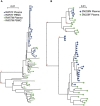
The HIV-1 epidemic in sub-Saharan Africa is driven largely by heterosexual transmission of non-subtype B viruses, of which subtypes C and A are predominant. Previous studies of subtype B and subtype C transmission pairs have suggested that a single variant from the chronically infected partner can establish infection in their newly infected partner. However, in subtype A infected individuals from a sex worker cohort and subtype B individuals from STD clinics, infection was frequently established by multiple variants. This study examined over 1750 single-genome amplified viral sequences derived from epidemiologically linked subtype C and subtype A transmission pairs very early after infection. In 90% (18/20) of the pairs, HIV-1 infection is initiated by a single viral variant that is derived from the quasispecies of the transmitting partner. In addition, the virus initiating infection in individuals who were infected by someone other than their spouse was characterized to determine if genital infections mitigated the severe genetic bottleneck observed in a majority of epidemiologically linked heterosexual HIV-1 transmission events. In nearly 50% (3/7) of individuals infected by someone other than their spouse, multiple genetic variants from a single individual established infection. A statistically significant association was observed between infection by multiple genetic variants and an inflammatory genital infection in the newly infected individual. Thus, in the vast majority of HIV-1 transmission events in cohabiting heterosexual couples, a single genetic variant establishes infection. Nevertheless, this severe genetic bottleneck can be mitigated by the presence of inflammatory genital infections in the at risk partner, suggesting that this restriction on genetic diversity is imposed in large part by the mucosal barrier.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- UNAIDS/WHO. AIDS Epidemic Update: 2007. AIDS Epidemic Updates. Geneva: The Joint United Nations Programme on HIV/AIDS and the World Health Organization; 2007.
-
- Zhu T, Mo H, Wang N, Nam DS, Cao Y, et al. Genotypic and phenotypic characterization of HIV-1 patients with primary infection. Science. 1993;261:1179–1181. - PubMed
-
- Long EM, Martin HL, Jr, Kreiss JK, Rainwater SM, Lavreys L, et al. Gender differences in HIV-1 diversity at time of infection. Nat Med. 2000;6:71–75. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

