Interplay of Mre11 nuclease with Dna2 plus Sgs1 in Rad51-dependent recombinational repair
- PMID: 19165339
- PMCID: PMC2625443
- DOI: 10.1371/journal.pone.0004267
Interplay of Mre11 nuclease with Dna2 plus Sgs1 in Rad51-dependent recombinational repair
Abstract
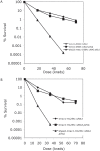
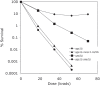
The Mre11/Rad50/Xrs2 complex initiates IR repair by binding to the end of a double-strand break, resulting in 5' to 3' exonuclease degradation creating a single-stranded 3' overhang competent for strand invasion into the unbroken chromosome. The nuclease(s) involved are not well understood. Mre11 encodes a nuclease, but it has 3' to 5', rather than 5' to 3' activity. Furthermore, mutations that inactivate only the nuclease activity of Mre11 but not its other repair functions, mre11-D56N and mre11-H125N, are resistant to IR. This suggests that another nuclease can catalyze 5' to 3' degradation. One candidate nuclease that has not been tested to date because it is encoded by an essential gene is the Dna2 helicase/nuclease. We recently reported the ability to suppress the lethality of a dna2Delta with a pif1Delta. The dna2Delta pif1Delta mutant is IR-resistant. We have determined that dna2Delta pif1Delta mre11-D56N and dna2Delta pif1Delta mre11-H125N strains are equally as sensitive to IR as mre11Delta strains, suggesting that in the absence of Dna2, Mre11 nuclease carries out repair. The dna2Delta pif1Delta mre11-D56N triple mutant is complemented by plasmids expressing Mre11, Dna2 or dna2K1080E, a mutant with defective helicase and functional nuclease, demonstrating that the nuclease of Dna2 compensates for the absence of Mre11 nuclease in IR repair, presumably in 5' to 3' degradation at DSB ends. We further show that sgs1Delta mre11-H125N, but not sgs1Delta, is very sensitive to IR, implicating the Sgs1 helicase in the Dna2-mediated pathway.
Conflict of interest statement
Figures





References
-
- Lisby M, Barlow JH, Burgess RC, Rothstein R. Choreography of the DNA damage response: Spatiotemporal relationships among checkpoint and repair proteins. Cell. 2004;118:699–713. - PubMed
-
- Sung P. Function of yeast Rad52 protein as a mediator between replication protein A and the Rad51 recombinase. J Biol Chem. 1997;272:28194–28197. - PubMed
-
- D'Amours D, Jackson SP. The Mre11 complex: at the crossroads of dna repair and checkpoint signalling. Nat Rev Mol Cell Biol. 2002;3:317–327. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

