Re-engineering a beta-lactamase using prototype peptides from a library of local structural motifs
- PMID: 19165724
- PMCID: PMC2708064
- DOI: 10.1002/pro.47
Re-engineering a beta-lactamase using prototype peptides from a library of local structural motifs
Abstract
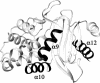
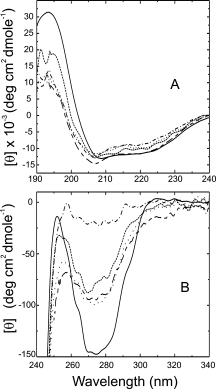
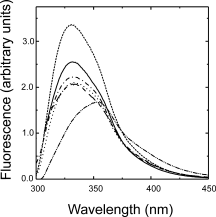
B. licheniformis exo-small beta-lactamase (ESBL) has a complex architecture with twelve alpha helices and a five-stranded beta sheet. We replaced, separately or simultaneously, three of the ESBL alpha helices with prototype amphiphatic helices from a catalog of secondary structure elements. Although the substitutes bear no sequence similarity to the originals and pertain to unrelated protein families, all the engineered ESBL variants were found able to fold in native like structures with in vitro and in vivo enzymic activity. The triple substituted variant resembles a primitive protein, with folding defects such as a strong tendency to oligomerization and very low stability; however it mimics a non homologous recombinant abandoning the family sequence space while preserving fold. The results test protein folding and evolution theories.
Figures
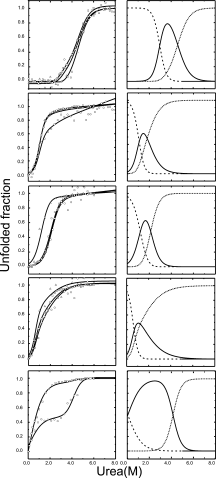
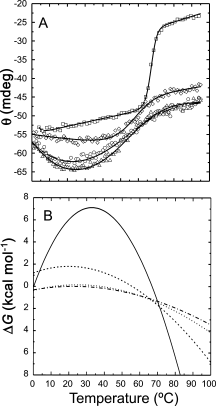
Similar articles
-
X-ray evidence of a native state with increased compactness populated by tryptophan-less B. licheniformis β-lactamase.Protein Sci. 2012 Jul;21(7):964-76. doi: 10.1002/pro.2076. Epub 2012 May 31. Protein Sci. 2012. PMID: 22496053 Free PMC article.
-
Export and folding of signal-sequenceless Bacillus licheniformis beta-lactamase in Escherichia coli.Eur J Biochem. 2000 Jun;267(12):3836-47. doi: 10.1046/j.1432-1327.2000.01422.x. Eur J Biochem. 2000. PMID: 10849003
-
Mapping the distribution of conformational information throughout a protein sequence.J Mol Biol. 2006 Apr 21;358(1):280-8. doi: 10.1016/j.jmb.2006.01.095. Epub 2006 Feb 9. J Mol Biol. 2006. PMID: 16510154
-
Increased folding stability of TEM-1 beta-lactamase by in vitro selection.J Mol Biol. 2008 Oct 31;383(1):238-51. doi: 10.1016/j.jmb.2008.07.082. Epub 2008 Aug 3. J Mol Biol. 2008. PMID: 18706424
-
Effect of disulfide-bond introduction on the activity and stability of the extended-spectrum class A beta-lactamase Toho-1.Biochim Biophys Acta. 2006 Aug;1764(8):1349-55. doi: 10.1016/j.bbapap.2006.06.004. Epub 2006 Jun 27. Biochim Biophys Acta. 2006. PMID: 16890032
Cited by
-
Fifty years of biophysics in Argentina.Biophys Rev. 2023 Aug 25;15(4):431-438. doi: 10.1007/s12551-023-01114-0. eCollection 2023 Aug. Biophys Rev. 2023. PMID: 37681102 Free PMC article.
-
Equilibrium partially folded states of B. licheniformis-lactamase.Eur Biophys J. 2019 May;48(4):341-348. doi: 10.1007/s00249-019-01361-8. Epub 2019 Mar 30. Eur Biophys J. 2019. PMID: 30929094
-
X-ray evidence of a native state with increased compactness populated by tryptophan-less B. licheniformis β-lactamase.Protein Sci. 2012 Jul;21(7):964-76. doi: 10.1002/pro.2076. Epub 2012 May 31. Protein Sci. 2012. PMID: 22496053 Free PMC article.
References
-
- Anfinsen CB. Principles that govern the folding of protein chains. Science. 1973;181:223–230. - PubMed
-
- Rennell D, Bouvier SE, Hardy LW, Poteete AR. Systematic mutation of bacteriophage T4 lysozyme. J Mol Biol. 1991;222:67–88. - PubMed
-
- Palzkill T, Botstein D. Probing β-lactamase structure and function using random replacement mutagenesis. Proteins. 1992;14:29–44. - PubMed
-
- Matthews BW. Structural and genetic analysis of protein stability. Annu Rev Biochem. 1993;62:139–160. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

