Transcriptomic comparison of Aspergillus niger growing on two different sugars reveals coordinated regulation of the secretory pathway
- PMID: 19166577
- PMCID: PMC2639373
- DOI: 10.1186/1471-2164-10-44
Transcriptomic comparison of Aspergillus niger growing on two different sugars reveals coordinated regulation of the secretory pathway
Abstract
Background: The filamentous fungus, Aspergillus niger, responds to nutrient availability by modulating secretion of various substrate degrading hydrolases. This ability has made it an important organism in industrial production of secreted glycoproteins. The recent publication of the A. niger genome sequence and availability of microarrays allow high resolution studies of transcriptional regulation of basal cellular processes, like those of glycoprotein synthesis and secretion. It is known that the activities of certain secretory pathway enzymes involved N-glycosylation are elevated in response to carbon source induced secretion of the glycoprotein glucoamylase. We have investigated whether carbon source dependent enhancement of protein secretion can lead to upregulation of secretory pathway elements extending beyond those involved in N-glycosylation.
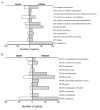
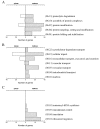
Results: This study compares the physiology and transcriptome of A. niger growing at the same specific growth rate (0.16 h(-1)) on xylose or maltose in carbon-limited chemostat cultures. Transcription profiles were obtained using Affymetrix GeneChip analysis of six replicate cultures for each of the two growth-limiting carbon sources. The production rate of extracellular proteins per gram dry mycelium was about three times higher on maltose compared to xylose. The defined culture conditions resulted in high reproducibility, discriminating even low-fold differences in transcription, which is characteristic of genes encoding basal cellular functions. This included elements in the secretory pathway and central metabolic pathways. Increased protein secretion on maltose was accompanied by induced transcription of > 90 genes related to protein secretion. The upregulated genes encode key elements in protein translocation to the endoplasmic reticulum (ER), folding, N-glycosylation, quality control, and vesicle packaging and transport between ER and Golgi. The induction effect of maltose resembles the unfolded protein response (UPR), which results from ER-stress and has previously been defined by treatment with chemicals interfering with folding of glycoproteins or by expression of heterologous proteins.
Conclusion: We show that upregulation of secretory pathway genes also occurs in conditions inducing secretion of endogenous glycoproteins - representing a more normal physiological state. Transcriptional regulation of protein synthesis and secretory pathway genes may thus reflect a general mechanism for modulation of secretion capacity in response to the conditional need for extracellular enzymes.
Figures

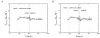

References
-
- Pel HJ, de Winde JH, Archer DB, Dyer PS, Hofmann G, Schaap PJ, Turner G, de Vries RP, Albang R, Albermann K, Andersen MR, Bendtsen JD, Benen JA, Berg M van den, Breestraat S, Caddick MX, Contreras R, Cornell M, Coutinho PM, Danchin EG, Debets AJ, Dekker P, van Dijck PW, van Dijk A, Dijkhuizen L, Driessen AJ, d'Enfert C, Geysens S, Goosen C, Groot GS, de Groot PW, Guillemette T, Henrissat B, Herweijer M, Hombergh JP van den, Hondel CA van den, Heijden RT van der, Kaaij RM van der, Klis FM, Kools HJ, Kubicek CP, van Kuyk PA, Lauber J, Lu X, Maarel MJ van der, Meulenberg R, Menke H, Mortimer MA, Nielsen J, Oliver SG, Olsthoorn M, Pal K, van Peij NN, Ram AF, Rinas U, Roubos JA, Sagt CM, Schmoll M, Sun J, Ussery D, Varga J, Vervecken W, Vondervoort PJ van de, Wedler H, Wösten HA, Zeng AP, van Ooyen AJ, Visser J, Stam H. Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nat Biotechnol. 2007;25:221–231. doi: 10.1038/nbt1282. - DOI - PubMed
-
- Pitt JI. Toxigenic fungi: which are important? Med Mycol. 2000;38:17–22. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

