An analysis of synteny of Arachis with Lotus and Medicago sheds new light on the structure, stability and evolution of legume genomes
- PMID: 19166586
- PMCID: PMC2656529
- DOI: 10.1186/1471-2164-10-45
An analysis of synteny of Arachis with Lotus and Medicago sheds new light on the structure, stability and evolution of legume genomes
Abstract
Background: Most agriculturally important legumes fall within two sub-clades of the Papilionoid legumes: the Phaseoloids and Galegoids, which diverged about 50 Mya. The Phaseoloids are mostly tropical and include crops such as common bean and soybean. The Galegoids are mostly temperate and include clover, fava bean and the model legumes Lotus and Medicago (both with substantially sequenced genomes). In contrast, peanut (Arachis hypogaea) falls in the Dalbergioid clade which is more basal in its divergence within the Papilionoids. The aim of this work was to integrate the genetic map of Arachis with Lotus and Medicago and improve our understanding of the Arachis genome and legume genomes in general. To do this we placed on the Arachis map, comparative anchor markers defined using a previously described bioinformatics pipeline. Also we investigated the possible role of transposons in the patterns of synteny that were observed.
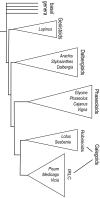
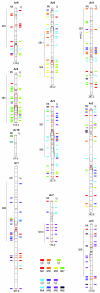
Results: The Arachis genetic map was substantially aligned with Lotus and Medicago with most synteny blocks presenting a single main affinity to each genome. This indicates that the last common whole genome duplication within the Papilionoid legumes predated the divergence of Arachis from the Galegoids and Phaseoloids sufficiently that the common ancestral genome was substantially diploidized. The Arachis and model legume genomes comparison made here, together with a previously published comparison of Lotus and Medicago allowed all possible Arachis-Lotus-Medicago species by species comparisons to be made and genome syntenies observed. Distinct conserved synteny blocks and non-conserved regions were present in all genome comparisons, implying that certain legume genomic regions are consistently more stable during evolution than others. We found that in Medicago and possibly also in Lotus, retrotransposons tend to be more frequent in the variable regions. Furthermore, while these variable regions generally have lower densities of single copy genes than the more conserved regions, some harbor high densities of the fast evolving disease resistance genes.
Conclusion: We suggest that gene space in Papilionoids may be divided into two broadly defined components: more conserved regions which tend to have low retrotransposon densities and are relatively stable during evolution; and variable regions that tend to have high retrotransposon densities, and whose frequent restructuring may fuel the evolution of some gene families.
Figures

References
-
- Lewis G, Schrire B, Mackinder B, Lock M, (Editors) Legumes of the World. Kew Publishing; 2005. ISBN 1 900 34780 6.
-
- FAOSTAT http://faostat.fao.org
-
- Kochert G, Stalker HT, Gimenes M, Galgaro L, Lopes CR, Moore K. RFLP and cytogenetic evidence on the origin and evolution of allotetraploid domesticated peanut, Arachis hypogaea (Leguminosae) Am J Bot. 1996;83:1282–1291. doi: 10.2307/2446112. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

