Patterns of exon-intron architecture variation of genes in eukaryotic genomes
- PMID: 19166620
- PMCID: PMC2636830
- DOI: 10.1186/1471-2164-10-47
Patterns of exon-intron architecture variation of genes in eukaryotic genomes
Abstract
Background: The origin and importance of exon-intron architecture comprises one of the remaining mysteries of gene evolution. Several studies have investigated the variations of intron length, GC content, ordinal position in a gene and divergence. However, there is little study about the structural variation of exons and introns.
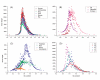
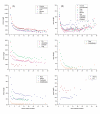
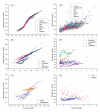
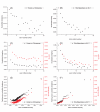
Results: We investigated the length, GC content, ordinal position and divergence in both exons and introns of 13 eukaryotic genomes, representing plant and animal. Our analyses revealed that three basic patterns of exon-intron variation were present in nearly all analyzed genomes (P < 0.001 in most cases): an ordinal reduction of length and divergence in both exon and intron, a co-variation between exon and its flanking introns in their length, GC content and divergence, and a decrease of average exon (or intron) length, GC content and divergence as the total exon numbers of a gene increased. In addition, we observed that the shorter introns had either low or high GC content, and the GC content of long introns was intermediate.
Conclusion: Although the factors contributing to these patterns have not been identified, our results provide three important clues: common factor(s) exist and may shape both exons and introns; the ordinal reduction patterns may reflect a time-orderly evolution; and the larger first and last exons may be splicing-required. These clues provide a framework for elucidating mechanisms involved in the organization of eukaryotic genomes and particularly in building exon-intron structures.
Figures
References
-
- Ohno S. So much "junk" DNA in our genome. Brookhaven Symp Biol. 1972;23:366–370. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

