Identification of a porcine DC-SIGN-related C-type lectin, porcine CLEC4G (LSECtin), and its order of intron removal during splicing: comparative genomic analyses of the cluster of genes CD23/CLEC4G/DC-SIGN among mammalian species
- PMID: 19166875
- PMCID: PMC7103215
- DOI: 10.1016/j.dci.2008.12.007
Identification of a porcine DC-SIGN-related C-type lectin, porcine CLEC4G (LSECtin), and its order of intron removal during splicing: comparative genomic analyses of the cluster of genes CD23/CLEC4G/DC-SIGN among mammalian species
Abstract
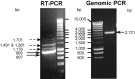
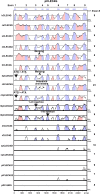
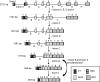
Human CLEC4G (previously named LSECtin), DC-SIGN, and L-SIGN are three important C-type lectins capable of mediating viral and bacterial pathogen recognitions. These three genes, together with CD23, form a lectin gene cluster at chromosome 19p13.3. In this study, we have experimentally identified the cDNA and the gene encoding porcine CLEC4G (pCLEC4G). Full-length pCLEC4G cDNA encodes a type II transmembrane protein of 290 amino acids. pCLEC4G gene has the same gene structure as the human and the predicted bovine, canis, mouse and rat CLEC4G genes with nine exons. A multi-species-conserved site at the extreme 3'-untranslated region of CLEC4G mRNAs was predicted to be targeted by microRNA miR-350 in domesticated animals and by miR-145 in primates, respectively. We detected pCLEC4G mRNA expression in liver, lymph node and spleen tissues. We also identified a series of sequential intermediate products of pCLEC4G pre-mRNA during splicing from pig liver. The previously unidentified porcine CD23 cDNA containing the complete coding region was subsequently cloned and found to express in spleen, thymus and lymph node. Furthermore, we compared the chromosomal regions syntenic to the human cluster of genes CD23/CLEC4G/DC-SIGN/L-SIGN in representative mammalian species including primates, domesticated animal, rodents and opossum. The L-SIGN homologues do not exist in non-primates mammals. The evolutionary processes of the gene cluster, from marsupials to primates, were proposed based upon their genomic structures and phylogenetic relationships.
Figures






Similar articles
-
Characterization of a novel C-type lectin-like gene, LSECtin: demonstration of carbohydrate binding and expression in sinusoidal endothelial cells of liver and lymph node.J Biol Chem. 2004 Apr 30;279(18):18748-58. doi: 10.1074/jbc.M311227200. Epub 2004 Jan 7. J Biol Chem. 2004. PMID: 14711836
-
Porcine DC-SIGN: molecular cloning, gene structure, tissue distribution and binding characteristics.Dev Comp Immunol. 2009 Apr;33(4):464-80. doi: 10.1016/j.dci.2008.09.010. Epub 2008 Oct 23. Dev Comp Immunol. 2009. PMID: 18951915 Free PMC article.
-
Five mouse homologues of the human dendritic cell C-type lectin, DC-SIGN.Int Immunol. 2001 Oct;13(10):1283-90. doi: 10.1093/intimm/13.10.1283. Int Immunol. 2001. PMID: 11581173
-
DC-SIGN, DC-SIGNR and LSECtin: C-type lectins for infection.Int Rev Immunol. 2014 Jan;33(1):54-66. doi: 10.3109/08830185.2013.834897. Epub 2013 Oct 24. Int Rev Immunol. 2014. PMID: 24156700 Review.
-
DC-SIGN (dendritic cell-specific ICAM-grabbing non-integrin) and DC-SIGN-related (DC-SIGNR): friend or foe?Clin Sci (Lond). 2003 Apr;104(4):437-46. Clin Sci (Lond). 2003. PMID: 12653690 Review.
Cited by
-
Characterization of a novel anti-human lymphocyte activation gene 3 (LAG-3) antibody for cancer immunotherapy.MAbs. 2019 Aug/Sep;11(6):1139-1148. doi: 10.1080/19420862.2019.1629239. Epub 2019 Jun 26. MAbs. 2019. PMID: 31242068 Free PMC article.
-
Tat peptide-mediated soluble expression of the membrane protein LSECtin-CRD in Escherichia coli.PLoS One. 2013 Dec 16;8(12):e83579. doi: 10.1371/journal.pone.0083579. eCollection 2013. PLoS One. 2013. PMID: 24358298 Free PMC article.
-
Characterization of the duplicate L-SIGN and DC-SIGN genes in miiuy croaker and evolutionary analysis of L-SIGN in fishes.Dev Comp Immunol. 2015 May;50(1):19-25. doi: 10.1016/j.dci.2015.01.004. Epub 2015 Jan 13. Dev Comp Immunol. 2015. PMID: 25596146 Free PMC article.
-
Characterization of the Immune Microenvironment and Identification of Biomarkers in Chronic Rhinosinusitis with Nasal Polyps Using Single-Cell RNA Sequencing and Transcriptome Analysis.J Inflamm Res. 2024 Jan 12;17:253-277. doi: 10.2147/JIR.S440409. eCollection 2024. J Inflamm Res. 2024. PMID: 38229690 Free PMC article.
-
The porcine innate immune system: an update.Dev Comp Immunol. 2014 Aug;45(2):321-43. doi: 10.1016/j.dci.2014.03.022. Epub 2014 Apr 4. Dev Comp Immunol. 2014. PMID: 24709051 Free PMC article. Review.
References
-
- Cambi A., Koopman M., Figdor C.G. How C-type lectins detect pathogens. Cell Microbiol. 2005;7(4):481–488. - PubMed
-
- Weis W.I., Taylor M.E., Drickamer K. The C-type lectin superfamily in the immune system. Immunol Rev. 1998;163:19–34. - PubMed
-
- Conrad D.H., Ford J.W., Sturgill J.L., Gibb D.R. CD23: an overlooked regulator of allergic disease. Curr Allergy Asthma Rep. 2007;7(5):331–337. - PubMed
-
- Geijtenbeek T.B., Krooshoop D.J., Bleijs D.A. DC-SIGN–ICAM-2 interaction mediates dendritic cell trafficking. Nat Immunol. 2000;1(4):353–357. - PubMed
-
- Geijtenbeek T.B., Torensma R., van Vliet S.J., van Duijnhoven G.C., Adema G.J., van Kooyk Y., Figdor C.G. Identification of DC-SIGN, a novel dendritic cell-specific ICAM-3 receptor that supports primary immune responses. Cell. 2000;100(5):575–585. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

