Model development for the viral Kcv potassium channel
- PMID: 19167299
- PMCID: PMC2716456
- DOI: 10.1016/j.bpj.2008.09.050
Model development for the viral Kcv potassium channel
Erratum in
- Biophys J. 2011 Jan 5;100(1):270
Abstract
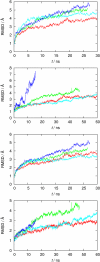
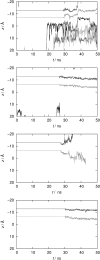
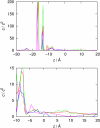
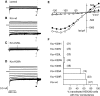
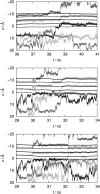
A computational model for the open state of the short viral Kcv potassium channel was created and tested based on homology modeling and extensive molecular-dynamics simulation in a membrane environment. Particular attention was paid to the structure of the highly flexible N-terminal region and to the protonation state of membrane-exposed lysine residues. Data from various experimental sources, NMR spectroscopy, and electrophysiology, as well as results from three-dimensional reference interaction site model integral equation theory were taken into account to select the most reasonable model among possible variants. The final model exhibits spontaneous ion transitions across the complete pore, with and without application of an external field. The nonequilibrium transport events could be induced reproducibly without abnormally large driving potential and without the need to place ions artificially at certain key positions along the transition path. The transport mechanism through the filter region corresponds to the classic view of single-file motion, which in our case is coupled to frequent exchange of ions between the innermost filter position and the cavity.
Figures


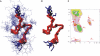

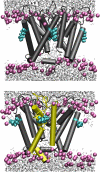

References
-
- Gazzarrini S., Severino M., Lombardi M., Morandi M., DiFrancesco D. The viral potassium channel Kcv: structural and functional features. FEBS Lett. 2003;552:12–16. - PubMed
-
- Plugge B., Gazzarrini S., Nelson M., Cerana R., Van Etten J.L. A potassium channel protein encoded by chlorella virus PBCV-1. Science. 2000;287:1641–1644. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

