Single-molecule fluorescence studies of a PH domain: new insights into the membrane docking reaction
- PMID: 19167305
- PMCID: PMC2716689
- DOI: 10.1016/j.bpj.2008.10.020
Single-molecule fluorescence studies of a PH domain: new insights into the membrane docking reaction
Abstract
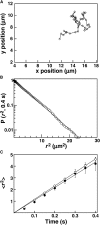
Proteins containing membrane targeting domains play essential roles in many cellular signaling pathways. However, important features of the membrane-bound state are invisible to bulk methods, thereby hindering mechanistic analysis of membrane targeting reactions. Here we use total internal reflection fluorescence microscopy (TIRFM), combined with single particle tracking, to probe the membrane docking mechanism of a representative pleckstrin homology (PH) domain isolated from the general receptor for phosphoinositides, isoform 1 (GRP1). The findings show three previously undescribed features of GRP1 PH domain docking to membranes containing its rare target lipid, phosphatidylinositol (3,4,5)-trisphosphate [PI(3,4,5)P(3)]. First, analysis of surface diffusion kinetics on supported lipid bilayers shows that in the absence of other anionic lipids, the PI(3,4,5)P(3)-bound protein exhibits the same diffusion constant as a single lipid molecule. Second, the binding of the anionic lipid phosphatidylserine to a previously unidentified secondary binding site slows both diffusion and dissociation kinetics. Third, TIRFM enables direct observation of rare events in which dissociation from the membrane surface is followed by transient diffusion through solution and rapid rebinding to a nearby, membrane-associated target lipid. Overall, this study shows that in vitro single-molecule TIRFM provides a new window into the molecular mechanisms of membrane docking reactions.
Figures





References
-
- Lemmon M.A. Membrane recognition by phospholipid-binding domains. Nat. Rev. Mol. Cell Biol. 2008;9:99–111. - PubMed
-
- Cho W., Stahelin R.V. Membrane-protein interactions in cell signaling and membrane trafficking. Annu. Rev. Biophys. Biomol. Struct. 2005;34:119–151. - PubMed
-
- DiNitto J.P., Cronin T.C., Lambright D.G. Membrane recognition and targeting by lipid-binding domains. Sci. STKE. 2003;2003:re16. - PubMed
-
- Jackson T.R., Kearns B.G., Theibert A.B. Cytohesins and centaurins: mediators of PI 3-kinase-regulated Arf signaling. Trends Biochem. Sci. 2000;25:489–495. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

