Epigenetic reprogramming and induced pluripotency
- PMID: 19168672
- PMCID: PMC2685952
- DOI: 10.1242/dev.020867
Epigenetic reprogramming and induced pluripotency
Abstract
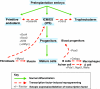
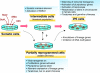
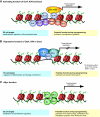
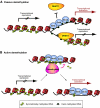
The cloning of animals from adult cells has demonstrated that the developmental state of adult cells can be reprogrammed into that of embryonic cells by uncharacterized factors within the oocyte. More recently, transcription factors have been identified that can induce pluripotency in somatic cells without the use of oocytes, generating induced pluripotent stem (iPS) cells. iPS cells provide a unique platform to dissect the molecular mechanisms that underlie epigenetic reprogramming. Moreover, iPS cells can teach us about principles of normal development and disease, and might ultimately facilitate the treatment of patients by custom-tailored cell therapy.
Figures

References
-
- Aasen, T., Raya, A., Barrero, M. J., Garreta, E., Consiglio, A., Gonzalez, F., Vassena, R., Bilic, J., Pekarik, V., Tiscornia, G. et al. (2008). Efficient and rapid generation of induced pluripotent stem cells from human keratinocytes. Nat. Biotechnol. 26, 1276-1284. - PubMed
-
- Aoi, T., Yae, K., Nakagawa, M., Ichisaka, T., Okita, K., Takahashi, K., Chiba, T. and Yamanaka, S. (2008). Generation of pluripotent stem cells from adult mouse liver and stomach cells. Science 321, 699-702. - PubMed
-
- Barreto, G., Schafer, A., Marhold, J., Stach, D., Swaminathan, S. K., Handa, V., Doderlein, G., Maltry, N., Wu, W., Lyko, F. et al. (2007). Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature 445, 671-675. - PubMed
-
- Bhattacharya, S. K., Ramchandani, S., Cervoni, N. and Szyf, M. (1999). A mammalian protein with specific demethylase activity for mCpG DNA. Nature 397, 579-583. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

