AgtA, the dicarboxylic amino acid transporter of Aspergillus nidulans, is concertedly down-regulated by exquisite sensitivity to nitrogen metabolite repression and ammonium-elicited endocytosis
- PMID: 19168757
- PMCID: PMC2653240
- DOI: 10.1128/EC.00270-08
AgtA, the dicarboxylic amino acid transporter of Aspergillus nidulans, is concertedly down-regulated by exquisite sensitivity to nitrogen metabolite repression and ammonium-elicited endocytosis
Abstract
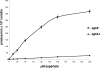
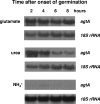
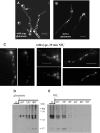
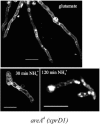
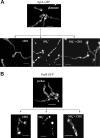
We identified agtA, a gene that encodes the specific dicarboxylic amino acid transporter of Aspergillus nidulans. The deletion of the gene resulted in loss of utilization of aspartate as a nitrogen source and of aspartate uptake, while not completely abolishing glutamate utilization. Kinetic constants showed that AgtA is a high-affinity dicarboxylic amino acid transporter and are in agreement with those determined for a cognate transporter activity identified previously. The gene is extremely sensitive to nitrogen metabolite repression, depends on AreA for its expression, and is seemingly independent from specific induction. We showed that the localization of AgtA in the plasma membrane necessitates the ShrA protein and that an active process elicited by ammonium results in internalization and targeting of AgtA to the vacuole, followed by degradation. Thus, nitrogen metabolite repression and ammonium-promoted vacuolar degradation act in concert to downregulate dicarboxylic amino acid transport activity.
Figures


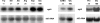


References
-
- Amillis, S., G. Cecchetto, V. Sophianopoulou, M. Koukaki, C. Scazzocchio, and G. Diallinas. 2004. Transcription of purine transporter genes is activated during the isotropic growth phase of Aspergillus nidulans conidia. Mol. Microbiol. 52205-216. - PubMed
-
- Andre, B. 1995. An overview of membrane transport proteins in Saccharomyces cerevisiae. Yeast 111575-1611. - PubMed
-
- Apostolaki, A. 2003. Topogenèse des transporteurs d'acides aminés chez le champignon filamenteux Aspergillus nidulans. Ph.D. thesis. Université Paris-Sud, Paris, France.
-
- Arst, H. N., Jr., and D. J. Cove. 1973. Nitrogen metabolite repression in Aspergillus nidulans. Mol. Gen. Genet. 126111-141. - PubMed
-
- Arst, H. N., Jr., S. A. Jones, and C. R. Bailey. 1981. A method for the selection of deletion mutations in the l-proline catabolism gene cluster of Aspergillus nidulans. Genet. Res. 38171-195. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

