Assigning strains to bacterial species via the internet
- PMID: 19171050
- PMCID: PMC2636762
- DOI: 10.1186/1741-7007-7-3
Assigning strains to bacterial species via the internet
Abstract
Background: Methods for assigning strains to bacterial species are cumbersome and no longer fit for purpose. The concatenated sequences of multiple house-keeping genes have been shown to be able to define and circumscribe bacterial species as sequence clusters. The advantage of this approach (multilocus sequence analysis; MLSA) is that, for any group of related species, a strain database can be produced and combined with software that allows query strains to be assigned to species via the internet. As an exemplar of this approach, we have studied a group of species, the viridans streptococci, which are very difficult to assign to species using standard taxonomic procedures, and have developed a website that allows species assignment via the internet.
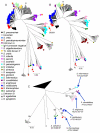
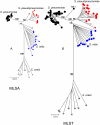
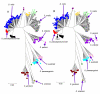
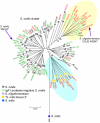
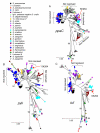
Results: Seven house-keeping gene sequences were obtained from 420 streptococcal strains to produce a viridans group database. The reference tree produced using the concatenated sequences identified sequence clusters which, by examining the position on the tree of the type strain of each viridans group species, could be equated with species clusters. MLSA also identified clusters that may correspond to new species, and previously described species whose status needs to be re-examined. A generic website and software for electronic taxonomy was developed. This site http://www.eMLSA.net allows the sequences of the seven gene fragments of a query strain to be entered and for the species assignment to be returned, according to its position within an assigned species cluster on the reference tree.
Conclusion: The MLSA approach resulted in the identification of well-resolved species clusters within this taxonomically challenging group and, using the software we have developed, allows unknown strains to be assigned to viridans species via the internet. Submission of new strains will provide a growing resource for the taxonomy of viridans group streptococci, allowing the recognition of potential new species and taxonomic anomalies. More generally, as the software at the MLSA website is generic, MLSA schemes and strain databases for other groups of related species can be hosted at this website, providing a portal for microbial electronic taxonomy.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

