Facile promoter deletion in Escherichia coli in response to leaky expression of very robust and benign proteins from common expression vectors
- PMID: 19171063
- PMCID: PMC2655282
- DOI: 10.1186/1475-2859-8-8
Facile promoter deletion in Escherichia coli in response to leaky expression of very robust and benign proteins from common expression vectors
Abstract
Background: Overexpression of proteins in Escherichia coli is considered routine today, at least when the protein is soluble and not otherwise toxic for the host. We report here that the massive overproduction of even such "benign" proteins can cause surprisingly efficient promoter deletions in the expression plasmid, leading to the growth of only non-producers, when expression is not well repressed in the newly transformed bacterial cell. Because deletion is so facile, it might impact on high-throughput protein production, e.g. for structural genomics, where not every expression parameter will be monitored.
Results: We studied the high-level expression of several robust non-toxic proteins using a T5 promoter under lac operator control. Full induction leads to no significant growth retardation. We compared expression from almost identical plasmids with or without the lacI gene together in strains expressing different levels of LacI. Any combination without net overexpression of LacI led to an efficient promoter deletion in the plasmid, although the number of growing colonies and even the plasmid size - all antibiotic-resistant non-producers - was almost normal, and thus the problem not immediately recognizable. However, by assuring sufficient repression during the initial establishment phase of the plasmid, deletion was completely prevented.
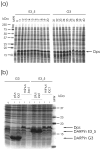
Conclusion: The deletions in the insufficiently repressed system are caused entirely by the burden of high-level translation. Since the E. coli Dps protein, known to protect DNA against stress in the stationary phase, is accumulated in the deletion mutants, the mutation may have taken place during a transient stationary phase. The cause of the deletion is thus distinct from the well known interference of high-level transcription with plasmid replication. The deletion can be entirely prevented by overexpressing LacI, a useful precaution even without any signs of stress caused by the protein.
Figures
Similar articles
-
Development of inducer-free expression plasmids based on IPTG-inducible promoters for Bacillus subtilis.Microb Cell Fact. 2017 Jul 25;16(1):130. doi: 10.1186/s12934-017-0747-0. Microb Cell Fact. 2017. PMID: 28743271 Free PMC article.
-
Escherichia coli σ70 promoters allow expression rate control at the cellular level in genome-integrated expression systems.Microb Cell Fact. 2020 Mar 5;19(1):58. doi: 10.1186/s12934-020-01311-6. Microb Cell Fact. 2020. PMID: 32138729 Free PMC article.
-
Using chromosomal lacIQ1 to control expression of genes on high-copy-number plasmids in Escherichia coli.Gene. 1998 Nov 26;223(1-2):221-31. doi: 10.1016/s0378-1119(98)00240-6. Gene. 1998. PMID: 9858738
-
Cooperative and anticooperative effects in binding of the first and second plasmid Osym operators to a LacI tetramer: evidence for contributions of non-operator DNA binding by wrapping and looping.J Mol Biol. 1996 Aug 2;260(5):697-717. doi: 10.1006/jmbi.1996.0431. J Mol Biol. 1996. PMID: 8709149
-
Use of high and low level overexpression plasmids to test mutant alleles of the recF gene of Escherichia coli K-12 for partial activity.Genetics. 1993 Nov;135(3):643-54. doi: 10.1093/genetics/135.3.643. Genetics. 1993. PMID: 8293970 Free PMC article.
Cited by
-
A New Synthetic Pathway for the Bioproduction of Glycolic Acid From Lignocellulosic Sugars Aimed at Maximal Carbon Conservation.Front Bioeng Biotechnol. 2019 Nov 27;7:359. doi: 10.3389/fbioe.2019.00359. eCollection 2019. Front Bioeng Biotechnol. 2019. PMID: 31850327 Free PMC article.
-
Jungle Express is a versatile repressor system for tight transcriptional control.Nat Commun. 2018 Sep 6;9(1):3617. doi: 10.1038/s41467-018-05857-3. Nat Commun. 2018. PMID: 30190458 Free PMC article.
-
Antagonism of PDGF-BB suppresses subretinal neovascularization and enhances the effects of blocking VEGF-A.Angiogenesis. 2014 Jul;17(3):553-62. doi: 10.1007/s10456-013-9402-5. Epub 2013 Oct 24. Angiogenesis. 2014. PMID: 24154861 Free PMC article.
-
Highly potent VEGF-A-antagonistic DARPins as anti-angiogenic agents for topical and intravitreal applications.Angiogenesis. 2013 Jan;16(1):101-11. doi: 10.1007/s10456-012-9302-0. Epub 2012 Sep 15. Angiogenesis. 2013. PMID: 22983424 Free PMC article.
-
Design and characterization of MP0250, a tri-specific anti-HGF/anti-VEGF DARPin® drug candidate.MAbs. 2017 Nov/Dec;9(8):1262-1269. doi: 10.1080/19420862.2017.1305529. MAbs. 2017. PMID: 29035637 Free PMC article.
References
-
- Merten O-W, Mattanovich D, Lang C, Larsson G, Neubauer P, Porro D, Postma P, Teixeira de Mattos J, Cole JAE. Recombinant protein production with prokaryotic and eukaryotic cells: A comparative view on host physiology. Springer Verlag; 2001.
-
- Glick BR. Metabolic load and heterologous gene expression. Biotechnol Adv. 1995;13:247–261. - PubMed
-
- Oh MK, Liao JC. DNA microarray detection of metabolic responses to protein overproduction in Escherichia coli. Metab Eng. 2000;2:201–209. - PubMed
-
- Andersson L, Yang S, Neubauer P, Enfors SO. Impact of plasmid presence and induction on cellular responses in fed batch cultures of Escherichia coli. J Biotechnol. 1996;46:255–263. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

