A consensus genetic map of sorghum that integrates multiple component maps and high-throughput Diversity Array Technology (DArT) markers
- PMID: 19171067
- PMCID: PMC2671505
- DOI: 10.1186/1471-2229-9-13
A consensus genetic map of sorghum that integrates multiple component maps and high-throughput Diversity Array Technology (DArT) markers
Abstract
Background: Sorghum genome mapping based on DNA markers began in the early 1990s and numerous genetic linkage maps of sorghum have been published in the last decade, based initially on RFLP markers with more recent maps including AFLPs and SSRs and very recently, Diversity Array Technology (DArT) markers. It is essential to integrate the rapidly growing body of genetic linkage data produced through DArT with the multiple genetic linkage maps for sorghum generated through other marker technologies. Here, we report on the colinearity of six independent sorghum component maps and on the integration of these component maps into a single reference resource that contains commonly utilized SSRs, AFLPs, and high-throughput DArT markers.
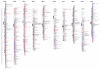
Results: The six component maps were constructed using the MultiPoint software. The lengths of the resulting maps varied between 910 and 1528 cM. The order of the 498 markers that segregated in more than one population was highly consistent between the six individual mapping data sets. The framework consensus map was constructed using a "Neighbours" approach and contained 251 integrated bridge markers on the 10 sorghum chromosomes spanning 1355.4 cM with an average density of one marker every 5.4 cM, and were used for the projection of the remaining markers. In total, the sorghum consensus map consisted of a total of 1997 markers mapped to 2029 unique loci (1190 DArT loci and 839 other loci) spanning 1603.5 cM and with an average marker density of 1 marker/0.79 cM. In addition, 35 multicopy markers were identified. On average, each chromosome on the consensus map contained 203 markers of which 58.6% were DArT markers. Non-random patterns of DNA marker distribution were observed, with some clear marker-dense regions and some marker-rare regions.
Conclusion: The final consensus map has allowed us to map a larger number of markers than possible in any individual map, to obtain a more complete coverage of the sorghum genome and to fill a number of gaps on individual maps. In addition to overall general consistency of marker order across individual component maps, good agreement in overall distances between common marker pairs across the component maps used in this study was determined, using a difference ratio calculation. The obtained consensus map can be used as a reference resource for genetic studies in different genetic backgrounds, in addition to providing a framework for transferring genetic information between different marker technologies and for integrating DArT markers with other genomic resources. DArT markers represent an affordable, high throughput marker system with great utility in molecular breeding programs, especially in crops such as sorghum where SNP arrays are not publicly available.
Figures


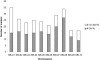
Similar articles
-
DArT markers: diversity analyses and mapping in Sorghum bicolor.BMC Genomics. 2008 Jan 22;9:26. doi: 10.1186/1471-2164-9-26. BMC Genomics. 2008. PMID: 18208620 Free PMC article.
-
A high-density consensus map of barley linking DArT markers to SSR, RFLP and STS loci and agricultural traits.BMC Genomics. 2006 Aug 12;7:206. doi: 10.1186/1471-2164-7-206. BMC Genomics. 2006. PMID: 16904008 Free PMC article.
-
A comprehensive genetic map of sugarcane that provides enhanced map coverage and integrates high-throughput Diversity Array Technology (DArT) markers.BMC Genomics. 2014 Feb 24;15(1):152. doi: 10.1186/1471-2164-15-152. BMC Genomics. 2014. PMID: 24564784 Free PMC article.
-
Computational approaches and software tools for genetic linkage map estimation in plants.Brief Bioinform. 2009 Nov;10(6):595-608. doi: 10.1093/bib/bbp045. Brief Bioinform. 2009. PMID: 19933208 Review.
-
Basic concepts and methodologies of DNA marker systems in plant molecular breeding.Heliyon. 2021 Sep 30;7(10):e08093. doi: 10.1016/j.heliyon.2021.e08093. eCollection 2021 Oct. Heliyon. 2021. PMID: 34765757 Free PMC article. Review.
Cited by
-
Drought stress tolerance strategies revealed by RNA-Seq in two sorghum genotypes with contrasting WUE.BMC Plant Biol. 2016 May 21;16(1):115. doi: 10.1186/s12870-016-0800-x. BMC Plant Biol. 2016. PMID: 27208977 Free PMC article.
-
Unraveling the genetic complexity underlying sorghum response to water availability.PLoS One. 2019 Apr 18;14(4):e0215515. doi: 10.1371/journal.pone.0215515. eCollection 2019. PLoS One. 2019. PMID: 30998785 Free PMC article.
-
Validation of QTL mapping and transcriptome profiling for identification of candidate genes associated with nitrogen stress tolerance in sorghum.BMC Plant Biol. 2017 Jul 11;17(1):123. doi: 10.1186/s12870-017-1064-9. BMC Plant Biol. 2017. PMID: 28697783 Free PMC article.
-
Two distinct classes of QTL determine rust resistance in sorghum.BMC Plant Biol. 2014 Dec 31;14:366. doi: 10.1186/s12870-014-0366-4. BMC Plant Biol. 2014. PMID: 25551674 Free PMC article.
-
Supermodels: sorghum and maize provide mutual insight into the genetics of flowering time.Theor Appl Genet. 2013 May;126(5):1377-95. doi: 10.1007/s00122-013-2059-z. Epub 2013 Mar 5. Theor Appl Genet. 2013. PMID: 23459955
References
-
- Kresovich S, Barbazuk B, Bedell JA, Borrell A, Buell CR, Burke J, Clifton S, Cordonnier-Pratt M, Cox S, Dahlberg J, Erpelding J, Fulton TM, Fulton B, Fulton L, Gingle AR, Hash CT, Huang Y, Jordan DR, Klein PE, Klein RR, Magalhaes J, McCombie R, Moore P, Mullet JE, Akins P, Paterson AH, Porter K, Pratt L, Roe B, Rooney W, Schnable PS, Stelly DM, Tuinstra M, Ware D, Warek U. Toward Sequencing the Sorghum Genome. A U.S. National Science Foundation-Sponsored Workshop Report. Plant Physiol. 2005;138:1898–1902. doi: 10.1104/pp.105.065136. - DOI - PMC - PubMed
-
- Klein RR, Mullet JE, Jordan DR, Miller FR, Rooney WL, Menz MA, Franks CD, Klein PE. The effect of tropical sorghum conversion and inbred development on genome diversity as revealed by high-resolution genotyping. Plant Genome. 2008. p. 1.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

