Lipidomic analysis of endocannabinoid metabolism in biological samples
- PMID: 19171504
- PMCID: PMC2723187
- DOI: 10.1016/j.jchromb.2009.01.008
Lipidomic analysis of endocannabinoid metabolism in biological samples
Erratum in
- J Chromatogr B Analyt Technol Biomed Life Sci. 2011 Jun 15;879(20):1844
Abstract
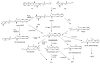
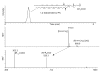
The endocannabinoids are signaling lipids present in many living organisms. They activate G protein-coupled cannabinoid receptors to modulate a broad range of biological processes that include emotion, cognition, inflammation and reproduction. The endocannabinoids are embedded in an interconnected network of cellular lipid pathways, the regulation of which is likely to control the strength and duration of endocannabinoid signals. Therefore, physiopathological or pharmacological perturbations of these pathways may indirectly affect endocannabinoid activity and, vice versa, endocannabinoid activity may influence lipid pathways involved in other metabolic and signaling events. Recent progress in liquid chromatography and mass spectrometry has fueled the development of targeted lipidomic approaches, which allow researchers to examine complex lipid interactions in cells and gain a broader view of the endocannabinoid system. Here, we review these new developments from the perspective of our laboratory's experience in the field.
Figures









References
-
- Katona I, Freund TF. Nat Med. 2008;14:923. - PubMed
-
- Di Marzo V. Nat Rev Drug Discov. 2008;7:438. - PubMed
-
- Piomelli D, Astarita G, Rapaka R. Nat Rev Neurosci. 2007;8:743. - PubMed
-
- Devane WA, Hanus L, Breuer A, Pertwee RG, Stevenson LA, Griffin G, Gibson D, Mandelbaum A, Etinger A, Mechoulam R. Science. 1992;258:1946. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R21 DA022702/DA/NIDA NIH HHS/United States
- R01 DA021644/DA/NIDA NIH HHS/United States
- R01DA-012447/DA/NIDA NIH HHS/United States
- R01 DA012447/DA/NIDA NIH HHS/United States
- P50 AG16573/AG/NIA NIH HHS/United States
- R21DA-022702/DA/NIDA NIH HHS/United States
- P50 AG016573/AG/NIA NIH HHS/United States
- RR274-305/3505998/RR/NCRR NIH HHS/United States
- R21 DA022478/DA/NIDA NIH HHS/United States
- RR274-297/3504008/RR/NCRR NIH HHS/United States
- 1RL1AA017538/AA/NIAAA NIH HHS/United States
- R01DK-073955/DK/NIDDK NIH HHS/United States
- R01 DA012413/DA/NIDA NIH HHS/United States
- R01 DK073955/DK/NIDDK NIH HHS/United States
- RL1 AA017538/AA/NIAAA NIH HHS/United States
- R01 DA-012413/DA/NIDA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

