A phosphomimetic mutation at threonine-57 abolishes transactivation activity and alters nuclear localization pattern of human pregnane x receptor
- PMID: 19171678
- PMCID: PMC2680541
- DOI: 10.1124/dmd.108.024695
A phosphomimetic mutation at threonine-57 abolishes transactivation activity and alters nuclear localization pattern of human pregnane x receptor
Abstract
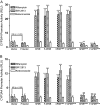
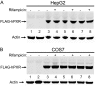
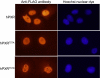
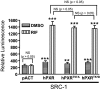
The pregnane X receptor (PXR) plays crucial roles in multiple physiological processes. However, the signaling mechanisms responsible are not well defined; it is most likely that multiple functions of PXR are modulated by its phosphorylation. Therefore, we sought to determine whether mutation at a highly conserved Thr(57) affects human PXR (hPXR) function. Site-directed mutagenesis was performed to generate phosphorylation-deficient (hPXR(T57A)) and phosphomimetic (hPXR(T57D)) mutants. Gene reporter, Western blotting, immunocytochemistry, mammalian two-hybrid, and electrophoretic mobility shift assays were used to study cytochrome P450 3A4 (CYP3A4) promoter activation, protein levels, localization, cofactor interaction, and CYP3A4 promoter binding of the hPXR mutants, respectively. hPXR(T57D), but not hPXR(T57A), lost its transcriptional activity. Neither mutation altered hPXR's protein levels and interaction with steroid receptor coactivator-1. hPXR and hPXR(T57A) exhibited a homogenous nuclear distribution, whereas hPXR(T57D) exhibited a distinctive punctate nuclear localization pattern similar to that of hPXR mutants with impaired function that colocalize with silencing mediator of retinoid and thyroid receptors (SMRT), although silencing of SMRT did not rescue the altered function of hPXR(T57D). However, hPXR(T57D), but not hPXR(T57A), impaired hPXR's ability to bind to the CYP3A4 promoter, consistent with the mutant's transactivation function. Furthermore, the 70-kDa form of ribosomal protein S6 kinase (p70 S6K) phosphorylated hPXR in vitro and inhibited its transcriptional activity, whereas hPXR(T57A) partially resisted the inhibitory effect of p70 S6K. Our studies identify a functionally significant phosphomimetic mutant (hPXR(T57D)) and show p70 S6K phosphorylation and regulation of hPXR transactivation to support the notion that phosphorylation plays important roles in regulating hPXR function.
Figures




Similar articles
-
Serine 350 of human pregnane X receptor is crucial for its heterodimerization with retinoid X receptor alpha and transactivation of target genes in vitro and in vivo.Biochem Pharmacol. 2015 Aug 15;96(4):357-68. doi: 10.1016/j.bcp.2015.06.018. Epub 2015 Jun 25. Biochem Pharmacol. 2015. PMID: 26119819 Free PMC article.
-
Protein phosphatase 2Cbetal regulates human pregnane X receptor-mediated CYP3A4 gene expression in HepG2 liver carcinoma cells.Drug Metab Dispos. 2010 Sep;38(9):1411-6. doi: 10.1124/dmd.110.032128. Epub 2010 Jun 10. Drug Metab Dispos. 2010. PMID: 20538721 Free PMC article.
-
Pregnane X receptor polymorphism affects CYP3A4 induction via a ligand-dependent interaction with steroid receptor coactivator-1.Pharmacogenet Genomics. 2007 May;17(5):369-82. doi: 10.1097/FPC.0b013e32803e40d7. Pharmacogenet Genomics. 2007. PMID: 17429319
-
Functional characterization of four naturally occurring variants of human pregnane X receptor (PXR): one variant causes dramatic loss of both DNA binding activity and the transactivation of the CYP3A4 promoter/enhancer region.Drug Metab Dispos. 2004 Jan;32(1):149-54. doi: 10.1124/dmd.32.1.149. Drug Metab Dispos. 2004. PMID: 14709632
-
Pregnane X receptor- and CYP3A4-humanized mouse models and their applications.Br J Pharmacol. 2011 Jun;163(3):461-8. doi: 10.1111/j.1476-5381.2010.01129.x. Br J Pharmacol. 2011. PMID: 21091656 Free PMC article. Review.
Cited by
-
Small-molecule modulators of PXR and CAR.Biochim Biophys Acta. 2016 Sep;1859(9):1141-1154. doi: 10.1016/j.bbagrm.2016.02.013. Epub 2016 Feb 24. Biochim Biophys Acta. 2016. PMID: 26921498 Free PMC article. Review.
-
Diindolylmethane, a naturally occurring compound, induces CYP3A4 and MDR1 gene expression by activating human PXR.Toxicol Lett. 2015 Feb 3;232(3):580-9. doi: 10.1016/j.toxlet.2014.12.015. Epub 2014 Dec 24. Toxicol Lett. 2015. PMID: 25542144 Free PMC article.
-
Overcoming drug resistance by regulating nuclear receptors.Adv Drug Deliv Rev. 2010 Oct 30;62(13):1257-64. doi: 10.1016/j.addr.2010.07.008. Epub 2010 Aug 4. Adv Drug Deliv Rev. 2010. PMID: 20691230 Free PMC article. Review.
-
Mutation of a single amino acid of pregnane X receptor switches an antagonist to agonist by altering AF-2 helix positioning.Cell Mol Life Sci. 2021 Jan;78(1):317-335. doi: 10.1007/s00018-020-03505-y. Epub 2020 Mar 30. Cell Mol Life Sci. 2021. PMID: 32232515 Free PMC article.
-
Serine 350 of human pregnane X receptor is crucial for its heterodimerization with retinoid X receptor alpha and transactivation of target genes in vitro and in vivo.Biochem Pharmacol. 2015 Aug 15;96(4):357-68. doi: 10.1016/j.bcp.2015.06.018. Epub 2015 Jun 25. Biochem Pharmacol. 2015. PMID: 26119819 Free PMC article.
References
-
- Bauer B, Yang X, Hartz AM, Olson ER, Zhao R, Kalvass JC, Pollack GM, and Miller DS (2006) In vivo activation of human pregnane X receptor tightens the blood-brain barrier to methadone through P-glycoprotein up-regulation. Mol Pharmacol 70 1212–1219. - PubMed
-
- Bhalla S, Ozalp C, Fang S, Xiang L, and Kemper JK (2004) Ligand-activated pregnane X receptor interferes with HNF-4 signaling by targeting a common coactivator PGC-1alpha. Functional implications in hepatic cholesterol and glucose metabolism. J Biol Chem 279 45139–45147. - PubMed
-
- Chen Y, Tang Y, Wang MT, Zeng S, and Nie D (2007) Human pregnane X receptor and resistance to chemotherapy in prostate cancer. Cancer Res 67 10361–10367. - PubMed
-
- Cohen P (2000) The regulation of protein function by multisite phosphorylation—a 25 year update. Trends Biochem Sci 25 596–601. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

