Arabidopsis ORC1 is a PHD-containing H3K4me3 effector that regulates transcription
- PMID: 19171893
- PMCID: PMC2644164
- DOI: 10.1073/pnas.0811093106
Arabidopsis ORC1 is a PHD-containing H3K4me3 effector that regulates transcription
Abstract
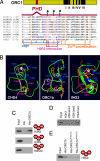
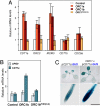
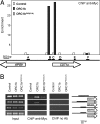
Control of gene expression depends on a complex and delicate balance of various posttranslational modifications of histones. However, the relevance of specific combinations of histone modifications is not fully defined. Downstream effector proteins recognize particular histone modifications and transduce this information into gene expression patterns. Methylation of histone H3 at lysine 4 (H3K4me) is a landmark of gene expression control in eukaryotes. Its recognition depends on the presence in the effector protein of a motif termed plant homeodomain (PHD) that specifically binds to H3K4me3. Here, we establish that Arabidopsis ORC1, the large subunit of the origin recognition complex involved in defining origins of DNA replication, functions as a transcriptional activator of a subset of genes, the promoters of which are preferentially bound by ORC1. Arabidopsis ORC1 contains a PHD and binds to H3K4me3. In addition to H4 acetylation, ORC1 binding correlates with increased H4K20me3 in the proximal promoter region of ORC1 targets. This suggests that H4K20me3, unlike in animal cells, is associated with transcriptional activation in Arabidopsis. Thus, our data provide a molecular basis for the opposite role of ORC1 in transcriptional activation in plants and repression in animals. Since only ORC1 proteins of plant species contain a PHD, we propose that plant ORC1 constitutes a novel class of H3K4me3 effector proteins characteristic of the plant kingdom.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. - PubMed
-
- Wysocka J, et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. - PubMed
-
- Sims RJ, Reinberg D. Histone H3 Lys 4 methylation: Caught in a bind? Genes Dev. 2006;20:2779–2786. - PubMed
-
- Fischer A, Hofmann I, Naumann K, Reuter G. Heterochromatin proteins and the control of heterochromatic gene silencing in Arabidopsis. J Plant Physiol. 2006;163:358–368. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

