The glutathione system of Aspergillus nidulans involves a fungus-specific glutathione S-transferase
- PMID: 19171936
- PMCID: PMC2658098
- DOI: 10.1074/jbc.M807771200
The glutathione system of Aspergillus nidulans involves a fungus-specific glutathione S-transferase
Abstract
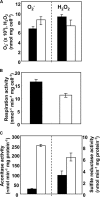
The tripeptide glutathione is involved in cellular defense mechanisms for xenobiotics and reactive oxygen species. This study investigated glutathione-dependent mechanisms in the model organism Aspergillus nidulans. A recombinant dimeric protein of A. nidulans glutathione reductase (GR) contained FAD and reduced oxidized glutathione (GSSG) using NADPH as an electron donor. A deletion strain of the GR gene (glrA) accumulated less intracellular reduced glutathione (GSH), indicating that the fungal GR contributes to GSSG reduction in vivo. Growth of the deletion strain of glrA was temperature-sensitive, and this phenotype was suppressed by adding GSH to the medium. The strain subsequently accumulated more intracellular superoxide, and cell-free respiration activity was partly defective. Growth of the strain decreased in the presence of oxidants, which induced glrA expression 1.5-6-fold. These results indicated that the fungal glutathione system functions as an antioxidant mechanism in A. nidulans. Our findings further revealed an initial proteomic differential display on GR-depleted and wild type strains. Up-regulation of thioredoxin reductase, peroxiredoxins, catalases, and cytochrome c peroxidase in the glrA-deletion strain revealed interplay between the glutathione system and both the thioredoxin system and hydrogen peroxide defense mechanisms. We also identified a hypothetical, up-regulated protein in the GR-depleted strains as glutathione S-transferase, which is unique among Ascomycetes fungi.
Figures




Similar articles
-
The DUG Pathway Governs Degradation of Intracellular Glutathione in Aspergillus nidulans.Appl Environ Microbiol. 2021 Apr 13;87(9):e01321-20. doi: 10.1128/AEM.01321-20. Print 2021 Apr 13. Appl Environ Microbiol. 2021. PMID: 33637571 Free PMC article.
-
Study on the glutathione metabolism of the filamentous fungus Aspergillus nidulans.Acta Microbiol Immunol Hung. 2017 Sep 1;64(3):255-272. doi: 10.1556/030.64.2017.003. Epub 2017 Mar 6. Acta Microbiol Immunol Hung. 2017. PMID: 28263103
-
Disruption of a glutathione reductase encoding gene in Acremonium chrysogenum leads to reduction of its growth, cephalosporin production and antioxidative ability which is recovered by exogenous methionine.Fungal Genet Biol. 2012 Feb;49(2):114-22. doi: 10.1016/j.fgb.2011.12.004. Epub 2011 Dec 19. Fungal Genet Biol. 2012. PMID: 22202809
-
The thioredoxin antioxidant system.Free Radic Biol Med. 2014 Jan;66:75-87. doi: 10.1016/j.freeradbiomed.2013.07.036. Epub 2013 Jul 27. Free Radic Biol Med. 2014. PMID: 23899494 Review.
-
Lighting the light reactions of photosynthesis by means of redox-responsive genetically encoded biosensors for photosynthetic intermediates.Photochem Photobiol Sci. 2023 Aug;22(8):2005-2018. doi: 10.1007/s43630-023-00425-1. Epub 2023 May 17. Photochem Photobiol Sci. 2023. PMID: 37195389 Review.
Cited by
-
The DUG Pathway Governs Degradation of Intracellular Glutathione in Aspergillus nidulans.Appl Environ Microbiol. 2021 Apr 13;87(9):e01321-20. doi: 10.1128/AEM.01321-20. Print 2021 Apr 13. Appl Environ Microbiol. 2021. PMID: 33637571 Free PMC article.
-
A community-driven reconstruction of the Aspergillus niger metabolic network.Fungal Biol Biotechnol. 2018 Sep 26;5:16. doi: 10.1186/s40694-018-0060-7. eCollection 2018. Fungal Biol Biotechnol. 2018. PMID: 30275963 Free PMC article.
-
Biochemical and ecophysiological responses to manganese stress by ectomycorrhizal fungus Pisolithus tinctorius and in association with Eucalyptus grandis.Mycorrhiza. 2016 Jul;26(5):475-87. doi: 10.1007/s00572-016-0686-3. Epub 2016 Feb 10. Mycorrhiza. 2016. PMID: 26861483
-
Illumina identification of RsrA, a conserved C2H2 transcription factor coordinating the NapA mediated oxidative stress signaling pathway in Aspergillus.BMC Genomics. 2014 Nov 22;15(1):1011. doi: 10.1186/1471-2164-15-1011. BMC Genomics. 2014. PMID: 25416206 Free PMC article.
-
Aspergillus nidulans gfdB, Encoding the Hyperosmotic Stress Protein Glycerol-3-phosphate Dehydrogenase, Disrupts Osmoadaptation in Aspergillus wentii.J Fungi (Basel). 2024 Apr 16;10(4):291. doi: 10.3390/jof10040291. J Fungi (Basel). 2024. PMID: 38667962 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

