Tools for fungal proteomics: multifunctional neurospora vectors for gene replacement, protein expression and protein purification
- PMID: 19171944
- PMCID: PMC2674810
- DOI: 10.1534/genetics.108.098707
Tools for fungal proteomics: multifunctional neurospora vectors for gene replacement, protein expression and protein purification
Abstract
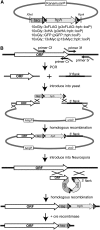
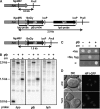
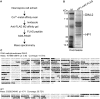
The completion of genome-sequencing projects for a number of fungi set the stage for detailed investigations of proteins. We report the generation of versatile expression vectors for detection and isolation of proteins and protein complexes in the filamentous fungus Neurospora crassa. The vectors, which can be adapted for other fungi, contain C- or N-terminal FLAG, HA, Myc, GFP, or HAT-FLAG epitope tags with a flexible poly-glycine linker and include sequences for targeting to the his-3 locus in Neurospora. To introduce mutations at native loci, we also made a series of knock-in vectors containing epitope tags followed by the selectable marker hph (resulting in hygromycin resistance) flanked by two loxP sites. We adapted the Cre/loxP system for Neurospora, allowing the selectable marker hph to be excised by introduction of Cre recombinase into a strain containing a knock-in cassette. Additionally, a protein purification method was developed on the basis of the HAT-FLAG tandem affinity tag system, which was used to purify HETEROCHROMATIN PROTEIN 1 (HP1) and associated proteins from Neurospora. As expected on the basis of yeast two-hybrid and co-immunoprecipitation (Co-IP) experiments, the Neurospora DNA methyltransferase DIM-2 was found in a complex with HP1. Features of the new vectors allowed for verification of an interaction between HP1 and DIM-2 in vivo by Co-IP assays on proteins expressed either from their native loci or from the his-3 locus.
Figures


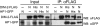
References
-
- Aebersold, R., and M. Mann, 2003. Mass spectrometry-based proteomics. Nature 422 198–207. - PubMed
-
- Bardiya, N., W. G. Alexander, T. D. Perdue, E. G. Barry, R. L. Metzenberg et al., 2008. Characterization of interactions between and among components of the meiotic silencing by unpaired DNA machinery in Neurospora crassa using bimolecular fluorescence complementation. Genetics 178 593–596. - PMC - PubMed
-
- Booher, K. R., and P. Kaiser, 2008. A PCR-based strategy to generate yeast strains expressing endogenous levels of amino-terminal epitope-tagged proteins. Biotech. J. 3 524–529. - PubMed
-
- Borjigin, J., and J. Nathans, 1994. Insertional mutagenesis as a probe of rhodopsin's topography, stability, and activity. J. Biol. Chem. 269 14715–14722. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

