Structure-function studies of the RNA polymerase II elongation complex
- PMID: 19171965
- PMCID: PMC2631633
- DOI: 10.1107/S0907444908039875
Structure-function studies of the RNA polymerase II elongation complex
Abstract
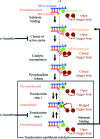
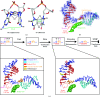
RNA polymerase II (Pol II) is the eukaryotic enzyme that is responsible for transcribing all protein-coding genes into messenger RNA (mRNA). The mRNA-transcription cycle can be divided into three stages: initiation, elongation and termination. During elongation, Pol II moves along a DNA template and synthesizes a complementary RNA chain in a processive manner. X-ray structural analysis has proved to be a potent tool for elucidating the mechanism of Pol II elongation. Crystallographic snapshots of different functional states of the Pol II elongation complex (EC) have elucidated mechanistic details of nucleotide addition and Pol II translocation. Further structural studies in combination with in vitro transcription experiments led to a mechanistic understanding of various additional features of the EC, including its inhibition by the fungal toxin alpha-amanitin, the tunability of the active site by the elongation factor TFIIS, the recognition of DNA lesions and the use of RNA as a template.
Figures




Similar articles
-
Structural basis of RNA polymerase II backtracking, arrest and reactivation.Nature. 2011 Mar 10;471(7337):249-53. doi: 10.1038/nature09785. Epub 2011 Feb 23. Nature. 2011. PMID: 21346759
-
RNA polymerase II structure: from core to functional complexes.Curr Opin Genet Dev. 2004 Apr;14(2):218-26. doi: 10.1016/j.gde.2004.01.003. Curr Opin Genet Dev. 2004. PMID: 15196470 Review.
-
The transcription elongation factor TFIIS is a component of RNA polymerase II preinitiation complexes.Proc Natl Acad Sci U S A. 2007 Oct 9;104(41):16068-73. doi: 10.1073/pnas.0704573104. Epub 2007 Oct 3. Proc Natl Acad Sci U S A. 2007. PMID: 17913884 Free PMC article.
-
Architecture of the RNA polymerase II-Paf1C-TFIIS transcription elongation complex.Nat Commun. 2017 Jun 6;8:15741. doi: 10.1038/ncomms15741. Nat Commun. 2017. PMID: 28585565 Free PMC article.
-
Architecture of the RNA polymerase II elongation complex: new insights into Spt4/5 and Elf1.Transcription. 2018;9(5):286-291. doi: 10.1080/21541264.2018.1454817. Epub 2018 May 7. Transcription. 2018. PMID: 29624124 Free PMC article. Review.
Cited by
-
The Human Isoform of RNA Polymerase II Subunit hRPB11bα Specifically Interacts with Transcription Factor ATF4.Int J Mol Sci. 2019 Dec 24;21(1):135. doi: 10.3390/ijms21010135. Int J Mol Sci. 2019. PMID: 31878192 Free PMC article.
-
T7 RNA polymerases backed up by covalently trapped proteins catalyze highly error prone transcription.J Biol Chem. 2012 Feb 24;287(9):6562-72. doi: 10.1074/jbc.M111.318410. Epub 2012 Jan 10. J Biol Chem. 2012. PMID: 22235136 Free PMC article.
-
RNA polymerase stalls in a post-translocated register and can hyper-translocate.Transcription. 2012 Sep-Oct;3(5):260-9. doi: 10.4161/trns.22307. Epub 2012 Sep 1. Transcription. 2012. PMID: 23132506 Free PMC article.
-
Macromolecular micromovements: how RNA polymerase translocates.Curr Opin Struct Biol. 2009 Dec;19(6):701-7. doi: 10.1016/j.sbi.2009.10.002. Epub 2009 Nov 2. Curr Opin Struct Biol. 2009. PMID: 19889534 Free PMC article. Review.
-
Soaking of DNA into crystals of archaeal RNA polymerase achieved by desalting in droplets.Acta Crystallogr Sect F Struct Biol Cryst Commun. 2012 Sep 1;68(Pt 9):1134-8. doi: 10.1107/S1744309112033507. Epub 2012 Aug 31. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2012. PMID: 22949213 Free PMC article.
References
-
- Armache, K.-J., Mitterweger, S., Meinhart, A. & Cramer, P. (2005). J. Biol. Chem.280, 7131–7134. - PubMed
-
- Broennimann, Ch., Eikenberry, E. F., Henrich, B., Horisberger, R., Huelsen, G., Pohl, E., Schmitt, B., Schulze-Briese, C., Suzuki, M., Tomizaki, T., Toyokawa, H. & Wagner, A. (2006). J. Synchrotron Rad.13, 120–130. - PubMed
-
- Brueckner, F. & Cramer, P. (2007). FEBS Lett.581, 2757–2760. - PubMed
-
- Brueckner, F. & Cramer, P. (2008). Nature Struct. Mol. Biol.15, 811–818. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

