Temporal analysis of neural differentiation using quantitative proteomics
- PMID: 19173612
- PMCID: PMC2693473
- DOI: 10.1021/pr8006667
Temporal analysis of neural differentiation using quantitative proteomics
Abstract
The ability to derive neural progenitors, differentiated neurons and glial cells from human embryonic stem cells (hESCs) with high efficiency holds promise for a number of clinical applications. However, investigating the temporal events is crucial for defining the underlying mechanisms that drive this process of differentiation along different lineages. We carried out quantitative proteomic profiling using a multiplexed approach capable of analyzing eight different samples simultaneously to monitor the temporal dynamics of protein abundance as human embryonic stem cells differentiate into motor neurons or astrocytes. With this approach, a catalog of approximately 1200 proteins along with their relative quantitative expression patterns was generated. The differential expression of the large majority of these proteins has not previously been reported or studied in the context of neural differentiation. As expected, two of the widely used markers of pluripotency, alkaline phosphatase (ALPL) and LIN28, were found to be downregulated during differentiation, while S-100 and tenascin C were upregulated in astrocytes. Neurofilament 3 protein, doublecortin and CAM kinase-like 1 and nestin proteins were upregulated during motor neuron differentiation. We identified a number of proteins whose expression was largely confined to specific cell types, embryonic stem cells, embryoid bodies and differentiating motor neurons. For example, glycogen phosphorylase (PYGL) and fatty acid binding protein 5 (FABP5) were enriched in ESCs, while beta spectrin (SPTBN5) was highly expressed in embryoid bodies. Karyopherin, heat shock 27 kDa protein 1 and cellular retinoic acid binding protein 2 (CRABP2) were upregulated in differentiating motor neurons but were downregulated in mature motor neurons. We validated some of the novel markers of the differentiation process using immunoblotting and immunocytochemical labeling. To our knowledge, this is the first large-scale temporal proteomic profiling of human stem cell differentiation into neural cell types highlighting proteins with limited or undefined roles in neural fate.
Figures

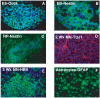

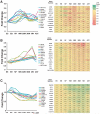
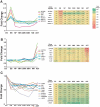

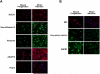
Similar articles
-
The cellular form of the prion protein guides the differentiation of human embryonic stem cells into neuron-, oligodendrocyte-, and astrocyte-committed lineages.Prion. 2014;8(3):266-75. doi: 10.4161/pri.32079. Epub 2014 Nov 1. Prion. 2014. PMID: 25486050 Free PMC article.
-
Quantitative proteomics analysis highlights the role of redox hemostasis and energy metabolism in human embryonic stem cell differentiation to neural cells.J Proteomics. 2014 Apr 14;101:1-16. doi: 10.1016/j.jprot.2014.02.002. Epub 2014 Feb 12. J Proteomics. 2014. PMID: 24530625
-
Regulation of mouse embryonic stem cell neural differentiation by retinoic acid.Dev Biol. 2009 Apr 15;328(2):456-71. doi: 10.1016/j.ydbio.2009.02.001. Epub 2009 Feb 13. Dev Biol. 2009. PMID: 19217899 Free PMC article.
-
Signals involved in neural differentiation of human embryonic stem cells.Neurosignals. 2009;17(4):234-41. doi: 10.1159/000231890. Epub 2009 Sep 30. Neurosignals. 2009. PMID: 19816060 Review.
-
Influence of Amino Acid Metabolism on Embryonic Stem Cell Function and Differentiation.Adv Nutr. 2016 Jul 15;7(4):780S-9S. doi: 10.3945/an.115.011031. Print 2016 Jul. Adv Nutr. 2016. PMID: 27422515 Free PMC article. Review.
Cited by
-
Neural Stem Cells (NSCs) and Proteomics.Mol Cell Proteomics. 2016 Feb;15(2):344-54. doi: 10.1074/mcp.O115.052704. Epub 2015 Oct 22. Mol Cell Proteomics. 2016. PMID: 26494823 Free PMC article. Review.
-
Neural differentiation in perspective: mitochondria as early programmers.Front Neurosci. 2025 Jan 8;18:1529855. doi: 10.3389/fnins.2024.1529855. eCollection 2024. Front Neurosci. 2025. PMID: 39844856 Free PMC article. Review.
-
PPARγ agonists promote oligodendrocyte differentiation of neural stem cells by modulating stemness and differentiation genes.PLoS One. 2012;7(11):e50500. doi: 10.1371/journal.pone.0050500. Epub 2012 Nov 21. PLoS One. 2012. PMID: 23185633 Free PMC article.
-
Proteomic analysis of stem cell differentiation and early development.Cold Spring Harb Perspect Biol. 2012 Mar 1;4(3):a008177. doi: 10.1101/cshperspect.a008177. Cold Spring Harb Perspect Biol. 2012. PMID: 22317846 Free PMC article. Review.
-
Effect of Ionizing Radiation on Transcriptome during Neural Differentiation of Human Embryonic Stem Cells.Radiat Res. 2020 May;193(5):460-470. doi: 10.1667/RR15535.1. Epub 2020 Mar 27. Radiat Res. 2020. PMID: 32216708 Free PMC article.
References
-
- Vallier L, Rugg-Gunn PJ, Bouhon IA, Andersson FK, Sadler AJ, Pedersen RA. Enhancing and diminishing gene function in human embryonic stem cells. Stem Cells. 2004;22(1):2–11. - PubMed
-
- Thomson JA. Embryonic stem cell lines derived from human blastocysts. 1998. [comment][erratum appears in Science 1998 Dec 4; 282(5395): 1827] - PubMed
-
- Reubinoff BE, Pera MF, Fong CY, Trounson A, Bongso A. Embryonic stem cell lines from human blastocysts: somatic differentiation in vitro. Nat Biotechnol. 2000;18(4):399–404. - PubMed
-
- Shin S, Sun Y, Liu Y, Khaner H, Svant S, Cai J, Xu QX, Davidson BP, Stice SL, Smith AK, Goldman SA, Reubinoff BE, Zhan M, Rao MS, Chesnut JD. Whole genome analysis of human neural stem cells derived from embryonic stem cells and stem and progenitor cells isolated from fetal tissue. Stem Cells. 2007;25(5):1298–306. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

