Ribosomal translocation: one step closer to the molecular mechanism
- PMID: 19173642
- PMCID: PMC3010847
- DOI: 10.1021/cb8002946
Ribosomal translocation: one step closer to the molecular mechanism
Abstract
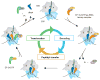
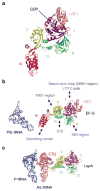
Protein synthesis occurs in ribosomes, the targets of numerous antibiotics. How these large and complex machines read and move along mRNA have proven to be challenging questions. In this Review, we focus on translocation, the last step of the elongation cycle in which movement of tRNA and mRNA is catalyzed by elongation factor G. Translocation entails large-scale movements of the tRNAs and conformational changes in the ribosome that require numerous tertiary contacts to be disrupted and reformed. We highlight recent progress toward elucidating the molecular basis of translocation and how various antibiotics influence tRNA-mRNA movement.
Figures



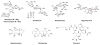
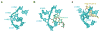

References
-
- Gromadski KB, Rodnina MV. Kinetic determinants of high-fidelity tRNA discrimination on the ribosome. Mol Cell. 2004;13:191–200. - PubMed
-
- Ogle JM, Ramakrishnan V. Structural insights into translational fidelity. Annu Rev Biochem. 2005;74:129–177. - PubMed
-
- Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science. 2000;289:905–920. - PubMed
-
- Carter AP, Clemons WM, Brodersen DE, Morgan-Warren RJ, Wimberly BT, Ramakrishnan V. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature. 2000;407:340–348. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

