Activation of mRNA translation by phage protein and low temperature: the case of Lactococcus lactis abortive infection system AbiD1
- PMID: 19173723
- PMCID: PMC2661086
- DOI: 10.1186/1471-2199-10-4
Activation of mRNA translation by phage protein and low temperature: the case of Lactococcus lactis abortive infection system AbiD1
Abstract
Background: Abortive infection (Abi) mechanisms comprise numerous strategies developed by bacteria to avoid being killed by bacteriophage (phage). Escherichia coli Abis are considered as mediators of programmed cell death, which is induced by infecting phage. Abis were also proposed to be stress response elements, but no environmental activation signals have yet been identified. Abis are widespread in Lactococcus lactis, but regulation of their expression remains an open question. We previously showed that development of AbiD1 abortive infection against phage bIL66 depends on orf1, which is expressed in mid-infection. However, molecular basis for this activation remains unclear.
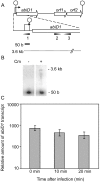
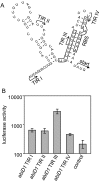
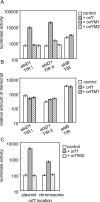
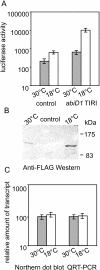
Results: In non-infected AbiD1+ cells, specific abiD1 mRNA is unstable and present in low amounts. It does not increase during abortive infection of sensitive phage. Protein synthesis directed by the abiD1 translation initiation region is also inefficient. The presence of the phage orf1 gene, but not its mutant AbiD1R allele, strongly increases abiD1 translation efficiency. Interestingly, cell growth at low temperature also activates translation of abiD1 mRNA and consequently the AbiD1 phenotype, and occurs independently of phage infection. There is no synergism between the two abiD1 inducers. Purified Orf1 protein binds mRNAs containing a secondary structure motif, identified within the translation initiation regions of abiD1, the mid-infection phage bIL66 M-operon, and the L. lactis osmC gene.
Conclusion: Expression of the abiD1 gene and consequently AbiD1 phenotype is specifically translationally activated by the phage Orf1 protein. The loss of ability to activate translation of abiD1 mRNA determines the molecular basis for phage resistance to AbiD1. We show for the first time that temperature downshift also activates abortive infection by activation of abiD1 mRNA translation.
Figures

Similar articles
-
Lactococcus lactis AbiD1 abortive infection efficiency is drastically increased by a phage protein.FEMS Microbiol Lett. 2002 Sep 10;214(2):283-7. doi: 10.1111/j.1574-6968.2002.tb11360.x. FEMS Microbiol Lett. 2002. PMID: 12351244
-
Phage operon involved in sensitivity to the Lactococcus lactis abortive infection mechanism AbiD1.J Bacteriol. 1995 Jul;177(13):3824-9. doi: 10.1128/jb.177.13.3824-3829.1995. J Bacteriol. 1995. PMID: 7601849 Free PMC article.
-
Characterization of the lactococcal abiD1 gene coding for phage abortive infection.J Bacteriol. 1995 Jul;177(13):3818-23. doi: 10.1128/jb.177.13.3818-3823.1995. J Bacteriol. 1995. PMID: 7601848 Free PMC article.
-
Phage abortive infection in lactococci: variations on a theme.Curr Opin Microbiol. 2005 Aug;8(4):473-9. doi: 10.1016/j.mib.2005.06.006. Curr Opin Microbiol. 2005. PMID: 15979388 Review.
-
Abortive infection antiphage defense systems: separating mechanism and phenotype.Trends Microbiol. 2023 Oct;31(10):1003-1012. doi: 10.1016/j.tim.2023.05.002. Epub 2023 May 31. Trends Microbiol. 2023. PMID: 37268559 Review.
Cited by
-
Combined Effects of Elevated pCO2 and Warming Facilitate Cyanophage Infections.Front Microbiol. 2017 Jun 13;8:1096. doi: 10.3389/fmicb.2017.01096. eCollection 2017. Front Microbiol. 2017. PMID: 28659906 Free PMC article.
-
Revenge of the phages: defeating bacterial defences.Nat Rev Microbiol. 2013 Oct;11(10):675-87. doi: 10.1038/nrmicro3096. Epub 2013 Aug 27. Nat Rev Microbiol. 2013. PMID: 23979432 Review.
-
A type IV modification-dependent restriction enzyme SauUSI from Staphylococcus aureus subsp. aureus USA300.Nucleic Acids Res. 2011 Jul;39(13):5597-610. doi: 10.1093/nar/gkr098. Epub 2011 Mar 17. Nucleic Acids Res. 2011. PMID: 21421560 Free PMC article.
-
The plasmid complement of Lactococcus lactis UC509.9 encodes multiple bacteriophage resistance systems.Appl Environ Microbiol. 2014 Jul;80(14):4341-9. doi: 10.1128/AEM.01070-14. Epub 2014 May 9. Appl Environ Microbiol. 2014. PMID: 24814781 Free PMC article.
-
Bacteriophage strategies for overcoming host antiviral immunity.Front Microbiol. 2023 Jun 8;14:1211793. doi: 10.3389/fmicb.2023.1211793. eCollection 2023. Front Microbiol. 2023. PMID: 37362940 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

