Independent saturation of three TrpRS subsites generates a partially assembled state similar to those observed in molecular simulations
- PMID: 19174517
- PMCID: PMC2644116
- DOI: 10.1073/pnas.0812752106
Independent saturation of three TrpRS subsites generates a partially assembled state similar to those observed in molecular simulations
Abstract
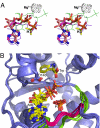
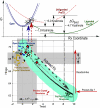
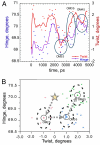
Two new crystal structures of Bacillus stearothermophilus tryptophanyl-tRNA synthetase (TrpRS) afford evidence that a closed interdomain hinge angle requires a covalent bond between AMP and an occupant of either pyrophosphate or tryptophan subsite. They also are within experimental error of a cluster of structures observed in a nonequilibrium molecular dynamics simulation showing partial active-site assembly. Further, the highest energy structure in a minimum action pathway computed by using elastic network models for Open and Pretransition state (PreTS) conformations for the fully liganded TrpRS monomer is intermediate between that simulated structure and a partially disassembled structure from a nonequilibrium molecular dynamics trajectory for the unliganded PreTS. These mutual consistencies provide unexpected validation of inferences drawn from molecular simulations.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Interconversion of ATP binding and conformational free energies by tryptophanyl-tRNA synthetase: structures of ATP bound to open and closed, pre-transition-state conformations.J Mol Biol. 2003 Jan 3;325(1):39-63. doi: 10.1016/s0022-2836(02)01156-7. J Mol Biol. 2003. PMID: 12473451
-
Computational studies of tryptophanyl-tRNA synthetase: activation of ATP by induced-fit.J Mol Biol. 2006 Oct 6;362(5):1159-80. doi: 10.1016/j.jmb.2006.06.078. Epub 2006 Sep 1. J Mol Biol. 2006. PMID: 16949606
-
Crystal structure of tryptophanyl-tRNA synthetase complexed with adenosine-5' tetraphosphate: evidence for distributed use of catalytic binding energy in amino acid activation by class I aminoacyl-tRNA synthetases.J Mol Biol. 2007 May 25;369(1):108-28. doi: 10.1016/j.jmb.2007.01.091. Epub 2007 Mar 12. J Mol Biol. 2007. PMID: 17428498 Free PMC article.
-
Substrate selection by aminoacyl-tRNA synthetases.Nucleic Acids Symp Ser. 1995;(33):40-2. Nucleic Acids Symp Ser. 1995. PMID: 8643392 Review.
-
Interdomain interactions in oligomeric enzymes: creation of asymmetry in homo-oligomers and role in metabolite channeling between active centers of hetero-oligomers.FEBS Lett. 2001 Jan 5;487(3):327-32. doi: 10.1016/s0014-5793(00)02338-3. FEBS Lett. 2001. PMID: 11163353 Review.
Cited by
-
Crystal structures of Saccharomyces cerevisiae tryptophanyl-tRNA synthetase: new insights into the mechanism of tryptophan activation and implications for anti-fungal drug design.Nucleic Acids Res. 2010 Jun;38(10):3399-413. doi: 10.1093/nar/gkp1254. Epub 2010 Jan 31. Nucleic Acids Res. 2010. PMID: 20123733 Free PMC article.
-
Combining multi-mutant and modular thermodynamic cycles to measure energetic coupling networks in enzyme catalysis.Struct Dyn. 2017 Jan 26;4(3):032101. doi: 10.1063/1.4974218. eCollection 2017 May. Struct Dyn. 2017. PMID: 28191480 Free PMC article.
-
Comparison of histidine recognition in human and trypanosomatid histidyl-tRNA synthetases.Biochimie. 2014 Nov;106:111-20. doi: 10.1016/j.biochi.2014.08.005. Epub 2014 Aug 20. Biochimie. 2014. PMID: 25151410 Free PMC article.
-
The mechanism of lineage-specific tRNA recognition by bacterial tryptophanyl-tRNA synthetase and its implications for inhibitor discovery.Nucleic Acids Res. 2025 May 22;53(10):gkaf466. doi: 10.1093/nar/gkaf466. Nucleic Acids Res. 2025. PMID: 40464690 Free PMC article.
-
Structural Robustness Affects the Engineerability of Aminoacyl-tRNA Synthetases for Genetic Code Expansion.Biochemistry. 2021 Feb 23;60(7):489-493. doi: 10.1021/acs.biochem.1c00056. Epub 2021 Feb 9. Biochemistry. 2021. PMID: 33560840 Free PMC article.
References
-
- Hammes-Schiffer S, Benkovic SJ. Relating protein motion to catalysis. Annu Rev Biochem. 2006;75:519–541. - PubMed
-
- Schiffer C, Hermans J. Promise of advances in simulation methods for protein crystallography: Implicit solvent models. Time averaging refinement, and quantum mechanical modeling. Methods Enzymol. 2003;374:412–461. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

