Improved tetracycline repressors for gene silencing in mycobacteria
- PMID: 19174563
- PMCID: PMC2665214
- DOI: 10.1093/nar/gkp015
Improved tetracycline repressors for gene silencing in mycobacteria
Abstract
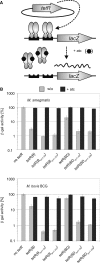
Tetracycline repressor (TetR)-controlled expression systems have recently been developed for mycobacteria and proven useful for the construction of conditional knockdown mutants and their analysis in vitro and during infections. However, even though these systems allowed tight regulation of some mycobacterial genes, they only showed limited or no phenotypic regulation for others. By adapting their codon usage to that of the Mycobacterium tuberculosis genome, we created tetR genes that mediate up to approximately 50-fold better repression of reporter gene activities in Mycobacterium smegmatis and Mycobacterium bovis BCG. In addition to these repressors, for which anhydrotetracycline (atc) functions as an inducer of gene expression, we used codon-usage adaption and structure-based design to develop improved reverse TetRs, for which atc functions as a corepressor. The previously described reverse repressor TetR only functioned when expressed from a strong promoter on a multicopy plasmid. The new reverse TetRs silence target genes more efficiently and allowed complete phenotypic silencing of M. smegmatis secA1 with chromosomally integrated tetR genes.
Figures
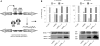


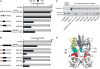


References
-
- Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE, III, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. - PubMed
-
- Sassetti CM, Boyd DH, Rubin EJ. Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 2003;48:77–84. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

