DNA mismatch binding and antiproliferative activity of rhodium metalloinsertors
- PMID: 19175313
- PMCID: PMC2747594
- DOI: 10.1021/ja8081044
DNA mismatch binding and antiproliferative activity of rhodium metalloinsertors
Abstract
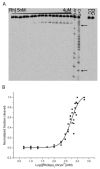
Deficiencies in mismatch repair (MMR) are associated with carcinogenesis. Rhodium metalloinsertors bind to DNA base mismatches with high specificity and inhibit cellular proliferation preferentially in MMR-deficient cells versus MMR-proficient cells. A family of chrysenequinone diimine complexes of rhodium with varying ancillary ligands that serve as DNA metalloinsertors has been synthesized, and both DNA mismatch binding affinities and antiproliferative activities against the human colorectal carcinoma cell lines HCT116N and HCT116O, an isogenic model system for MMR deficiency, have been determined. DNA photocleavage experiments reveal that all complexes bind to the mismatch sites with high specificities; DNA binding affinities to oligonucleotides containing single base CA and CC mismatches, obtained through photocleavage titration or competition, vary from 10(4) to 10(8) M(-1) for the series of complexes. Significantly, binding affinities are found to be inversely related to ancillary ligand size and directly related to differential inhibition of the HCT116 cell lines. The observed trend in binding affinity is consistent with the metalloinsertion mode where the complex binds from the minor groove with ejection of mismatched base pairs. The correlation between binding affinity and targeting of the MMR-deficient cell line suggests that rhodium metalloinsertors exert their selective biological effects on MMR-deficient cells through mismatch binding in vivo.
Figures





Similar articles
-
A Family of Rhodium Complexes with Selective Toxicity toward Mismatch Repair-Deficient Cancers.J Am Chem Soc. 2018 Apr 25;140(16):5612-5624. doi: 10.1021/jacs.8b02271. Epub 2018 Apr 17. J Am Chem Soc. 2018. PMID: 29620877 Free PMC article.
-
An unusual ligand coordination gives rise to a new family of rhodium metalloinsertors with improved selectivity and potency.J Am Chem Soc. 2014 Oct 8;136(40):14160-72. doi: 10.1021/ja5072064. Epub 2014 Sep 25. J Am Chem Soc. 2014. PMID: 25254630 Free PMC article.
-
Selective cytotoxicity of rhodium metalloinsertors in mismatch repair-deficient cells.Biochemistry. 2011 Dec 20;50(50):10919-28. doi: 10.1021/bi2015822. Epub 2011 Nov 21. Biochemistry. 2011. PMID: 22103240 Free PMC article.
-
Targeting DNA mismatches with metal complexes.J Inorg Biochem. 2025 Oct;271:112977. doi: 10.1016/j.jinorgbio.2025.112977. Epub 2025 Jun 24. J Inorg Biochem. 2025. PMID: 40592119 Review.
-
Detection of DNA base mismatches using DNA intercalators.Methods Enzymol. 2002;353:506-22. doi: 10.1016/s0076-6879(02)53073-1. Methods Enzymol. 2002. PMID: 12078523 Review. No abstract available.
Cited by
-
A bulky rhodium complex bound to an adenosine-adenosine DNA mismatch: general architecture of the metalloinsertion binding mode.Biochemistry. 2009 May 26;48(20):4247-53. doi: 10.1021/bi900194e. Biochemistry. 2009. PMID: 19374348 Free PMC article.
-
Rational guide RNA engineering for small-molecule control of CRISPR/Cas9 and gene editing.Nucleic Acids Res. 2022 May 6;50(8):4769-4783. doi: 10.1093/nar/gkac255. Nucleic Acids Res. 2022. PMID: 35446403 Free PMC article.
-
A potential rhodium cancer therapy: studies of a cytotoxic organorhodium(I) complex that binds DNA.Bioorg Med Chem Lett. 2013 May 1;23(9):2527-31. doi: 10.1016/j.bmcl.2013.03.016. Epub 2013 Mar 14. Bioorg Med Chem Lett. 2013. PMID: 23541673 Free PMC article.
-
Research Progress of Metal Anticancer Drugs.Pharmaceutics. 2023 Dec 11;15(12):2750. doi: 10.3390/pharmaceutics15122750. Pharmaceutics. 2023. PMID: 38140091 Free PMC article. Review.
-
Rhodium metalloinsertor binding generates a lesion with selective cytotoxicity for mismatch repair-deficient cells.Proc Natl Acad Sci U S A. 2017 Jul 3;114(27):6948-6953. doi: 10.1073/pnas.1706665114. Epub 2017 Jun 20. Proc Natl Acad Sci U S A. 2017. PMID: 28634291 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

