Systematic analysis of insertions and deletions specific to nematode proteins and their proposed functional and evolutionary relevance
- PMID: 19175938
- PMCID: PMC2644674
- DOI: 10.1186/1471-2148-9-23
Systematic analysis of insertions and deletions specific to nematode proteins and their proposed functional and evolutionary relevance
Abstract
Background: Amino acid insertions and deletions in proteins are considered relatively rare events, and their associations with the evolution and adaptation of organisms are not yet understood. In this study, we undertook a systematic analysis of over 214,000 polypeptides from 32 nematode species and identified insertions and deletions unique to nematode proteins in more than 1000 families and provided indirect evidence that these alterations are linked to the evolution and adaptation of nematodes.
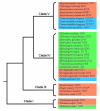
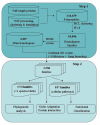
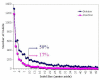
Results: Amino acid alterations in sequences of nematodes were identified by comparison with homologous sequences from a wide range of eukaryotic (metzoan) organisms. This comparison revealed that the proteins inferred from transcriptomic datasets for nematodes contained more deletions than insertions, and that the deletions tended to be larger in length than insertions, indicating a decreased size of the transcriptome of nematodes compared with other organisms. The present findings showed that this reduction is more pronounced in parasitic nematodes compared with the free-living nematodes of the genus Caenorhabditis. Consistent with a requirement for conservation in proteins involved in the processing of genetic information, fewer insertions and deletions were detected in such proteins. On the other hand, more insertions and deletions were recorded for proteins inferred to be involved in the endocrine and immune systems, suggesting a link with adaptation. Similarly, proteins involved in multiple cellular pathways tended to display more deletions and insertions than those involved in a single pathway. The number of insertions and deletions shared by a range of plant parasitic nematodes were higher for proteins involved in lipid metabolism and electron transport compared with other nematodes, suggesting an association between metabolic adaptation and parasitism in plant hosts. We also identified three sizable deletions from proteins found to be specific to and shared by parasitic nematodes, which, given their uniqueness, might serve as target candidates for drug design.
Conclusion: This study illustrates the significance of using comparative genomics approaches to identify molecular elements unique to parasitic nematodes, which have adapted to a particular host organism and mode of existence during evolution. While the focus of this study was on nematodes, the approach has applicability to a wide range of other groups of organisms.
Figures


Similar articles
-
Genomic Analyses Identify Novel Molecular Signatures Specific for the Caenorhabditis and other Nematode Taxa Providing Novel Means for Genetic and Biochemical Studies.Genes (Basel). 2019 Sep 24;10(10):739. doi: 10.3390/genes10100739. Genes (Basel). 2019. PMID: 31554175 Free PMC article.
-
Needles in the EST haystack: large-scale identification and analysis of excretory-secretory (ES) proteins in parasitic nematodes using expressed sequence tags (ESTs).PLoS Negl Trop Dis. 2008 Sep 24;2(9):e301. doi: 10.1371/journal.pntd.0000301. PLoS Negl Trop Dis. 2008. PMID: 18820748 Free PMC article.
-
Trichinella spiralis: Adaptation and parasitism.Vet Parasitol. 2016 Nov 15;231:8-21. doi: 10.1016/j.vetpar.2016.07.003. Epub 2016 Jul 2. Vet Parasitol. 2016. PMID: 27425574 Free PMC article.
-
Role of horizontal gene transfer in the evolution of plant parasitism among nematodes.Methods Mol Biol. 2009;532:517-35. doi: 10.1007/978-1-60327-853-9_30. Methods Mol Biol. 2009. PMID: 19271205 Review.
-
Caenorhabditis elegans is a nematode.Science. 1998 Dec 11;282(5396):2041-6. doi: 10.1126/science.282.5396.2041. Science. 1998. PMID: 9851921 Review.
Cited by
-
Genomic Analyses Identify Novel Molecular Signatures Specific for the Caenorhabditis and other Nematode Taxa Providing Novel Means for Genetic and Biochemical Studies.Genes (Basel). 2019 Sep 24;10(10):739. doi: 10.3390/genes10100739. Genes (Basel). 2019. PMID: 31554175 Free PMC article.
-
Pan-Nematoda Transcriptomic Elucidation of Essential Intestinal Functions and Therapeutic Targets With Broad Potential.EBioMedicine. 2015 Jul 29;2(9):1079-89. doi: 10.1016/j.ebiom.2015.07.030. eCollection 2015 Sep. EBioMedicine. 2015. PMID: 26501106 Free PMC article.
-
The draft genome of the parasitic nematode Trichinella spiralis.Nat Genet. 2011 Mar;43(3):228-35. doi: 10.1038/ng.769. Epub 2011 Feb 20. Nat Genet. 2011. PMID: 21336279 Free PMC article.
-
Phylogenetic and genetic evolutionary analyses of the mitochondrial genome of Mastophorus muris in Neodon fuscus from the Qinghai-Tibetan Plateau.Microbiol Spectr. 2025 Aug 5;13(8):e0306724. doi: 10.1128/spectrum.03067-24. Epub 2025 Jul 7. Microbiol Spectr. 2025. PMID: 40621925 Free PMC article.
-
Characterizing Ancylostoma caninum transcriptome and exploring nematode parasitic adaptation.BMC Genomics. 2010 May 14;11:307. doi: 10.1186/1471-2164-11-307. BMC Genomics. 2010. PMID: 20470405 Free PMC article.
References
-
- Barak Y, Nov Y, Ackerley DF, Matin A. Enzyme improvement in the absence of structural knowledge: a novel statistical approach. ISME Journal. 2007;2(2):171–179. - PubMed
-
- Gillam EMJ. Engineering Cytochrome P450 Enzymes. Chem Res Toxicol. 2008;21(1):220–231. - PubMed
-
- Kaur J, Sharma R. Directed evolution: An approach to engineer enzymes. Crit Rev Biotechnol. 2006;26(3):165–199. - PubMed
-
- Kazlauskas RJ. Enhancing catalytic promiscuity for biocatalysis. Curr Opin Chem Biol. 2005;9(2):195–201. - PubMed
-
- Siest G, Jeannesson E, Visvikis-Siest S. Enzymes and pharmacogenetics of cardiovascular drugs. Clin Chim Acta. 2007;381(1):26–31. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

