Molecular mechanisms of thioredoxin and glutaredoxin as hydrogen donors for Mammalian s phase ribonucleotide reductase
- PMID: 19176520
- PMCID: PMC2659180
- DOI: 10.1074/jbc.M809338200
Molecular mechanisms of thioredoxin and glutaredoxin as hydrogen donors for Mammalian s phase ribonucleotide reductase
Abstract
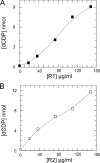
Ribonucleotide reductase (RNR) catalyzes the rate-limiting step in deoxyribonucleotide synthesis essential for DNA replication and repair. RNR in S phase mammalian cells comprises a weak cytosolic complex of the catalytic R1 protein containing redox active cysteine residues and the R2 protein harboring the tyrosine free radical. Each enzyme turnover generates a disulfide in the active site of R1, which is reduced by C-terminally located shuttle dithiols leaving a disulfide to be reduced. Electrons for reduction come ultimately from NADPH via thioredoxin reductase and thioredoxin (Trx) or glutathione reductase, glutathione, and glutaredoxin (Grx), but the mechanism has not been clarified for mammalian RNR. Using recombinant mouse RNR, we found that Trx1 and Grx1 had similar catalytic efficiency (k(cat)/K(m)). With 4 mm GSH, Grx1 showed a higher affinity (apparent K(m) value, 0.18 microm) compared with Trx1 which displayed a higher apparent k(cat), suggesting its major role in S phase DNA replication. Surprisingly, Grx activity was strongly dependent on GSH concentrations (apparent K(m) value, 3 mm) and a Grx2 C40S mutant was active despite only one cysteine residue in the active site. This demonstrates a GSH-mixed disulfide mechanism for glutaredoxin catalysis in contrast to the dithiol mechanism for thioredoxin. This may be an advantage with the low levels of RNR for DNA repair or in tumor cells with high RNR and no or low Trx expression. Our results demonstrate mechanistic differences between the mammalian and canonical Escherichia coli RNR enzymes, which may offer an explanation for the nonconserved shuttle dithiol sequences in the C terminus of the R1.
Figures






References
-
- Nordlund, P., and Reichard, P. (2006) Annu. Rev. Biochem. 75 681–706 - PubMed
-
- Kolberg, M., Strand, K. R., Graff, P., and Andersson, K. K. (2004) Biochim. Biophys. Acta 1699 1–34 - PubMed
-
- Reichard, P. (2002) Arch. Biochem. Biophys. 397 149–155 - PubMed
-
- Kashlan, O. B., Scott, C. P., Lear, J. D., and Cooperman, B. S. (2002) Biochemistry 41 462–474 - PubMed
-
- Rofougaran, R., Vodnala, M., and Hofer, A. (2006) J. Biol. Chem. 281 27705–27711 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

