Structural and functional analysis of transmembrane XI of the NHE1 isoform of the Na+/H+ exchanger
- PMID: 19176522
- PMCID: PMC2670159
- DOI: 10.1074/jbc.M809201200
Structural and functional analysis of transmembrane XI of the NHE1 isoform of the Na+/H+ exchanger
Abstract
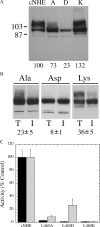
The Na(+)/H(+) exchanger isoform 1 is a ubiquitously expressed integral membrane protein that regulates intracellular pH in mammals by extruding an intracellular H(+) in exchange for one extracellular Na(+). We characterized structural and functional aspects of the critical transmembrane (TM) segment XI (residues 449-470) by using cysteine scanning mutagenesis and high resolution NMR. Each residue of TM XI was mutated to cysteine in the background of the cysteine-less protein and the sensitivity to water-soluble sulfhydryl reactive compounds MTSET ((2-(trimethylammonium) ethyl)methanethiosulfonate) and MTSES ((2-sulfonatoethyl) methanethiosulfonate) was determined for those residues with at least moderate activity remaining. Of the residues tested, only proteins with mutations L457C, I461C, and L465C were inhibited by MTSET. The activity of the L465C mutant was almost completely eliminated, whereas that of the L457C and I461C mutants was partially affected. The structure of a peptide representing TM XI (residues Lys(447)-Lys(472)) was determined using high resolution NMR spectroscopy in dodecylphosphocholine micelles. The structure consisted of helical regions between Asp(447)-Tyr(454) and Phe(460)-Lys(471) at the N and C termini of the peptide, respectively, connected by a region with poorly defined, irregular structure consisting of residues Gly(455)-Gly(459). TM XI of NHE1 had a structural similarity to TM XI of the Escherichia coli Na(+)/H(+) exchanger NhaA. The results suggest that TM XI is a discontinuous helix, with residue Leu(465) contributing to the pore.
Figures


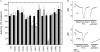

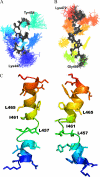

References
-
- Karmazyn, M., Sawyer, M., and Fliegel, L. (2005) Curr. Drug Targets Cardiovasc. Haematol. Disord. 5 323-335 - PubMed
-
- Grinstein, S., Rotin, D., and Mason, M. J. (1989) Biochim. Biophys. Acta 988 73-97 - PubMed
-
- Shrode, L., Cabado, A., Goss, G., and Grinstein, S. (1996) in The Na+/H+ Exchanger (Fliegel, R. G., ed) pp. 101-122, Landes Company, Austin, TX
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Miscellaneous

