Fragment-based identification of druggable 'hot spots' of proteins using Fourier domain correlation techniques
- PMID: 19176554
- PMCID: PMC2647826
- DOI: 10.1093/bioinformatics/btp036
Fragment-based identification of druggable 'hot spots' of proteins using Fourier domain correlation techniques
Abstract
Motivation: The binding sites of proteins generally contain smaller regions that provide major contributions to the binding free energy and hence are the prime targets in drug design. Screening libraries of fragment-sized compounds by NMR or X-ray crystallography demonstrates that such 'hot spot' regions bind a large variety of small organic molecules, and that a relatively high 'hit rate' is predictive of target sites that are likely to bind drug-like ligands with high affinity. Our goal is to determine the 'hot spots' computationally rather than experimentally.
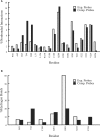
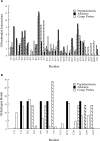
Results: We have developed the FTMAP algorithm that performs global search of the entire protein surface for regions that bind a number of small organic probe molecules. The search is based on the extremely efficient fast Fourier transform (FFT) correlation approach which can sample billions of probe positions on dense translational and rotational grids, but can use only sums of correlation functions for scoring and hence is generally restricted to very simple energy expressions. The novelty of FTMAP is that we were able to incorporate and represent on grids a detailed energy expression, resulting in a very accurate identification of low-energy probe clusters. Overlapping clusters of different probes are defined as consensus sites (CSs). We show that the largest CS is generally located at the most important subsite of the protein binding site, and the nearby smaller CSs identify other important subsites. Mapping results are presented for elastase whose structure has been solved in aqueous solutions of eight organic solvents, and we show that FTMAP provides very similar information. The second application is to renin, a long-standing pharmaceutical target for the treatment of hypertension, and we show that the major CSs trace out the shape of the first approved renin inhibitor, aliskiren.
Availability: FTMAP is available as a server at http://ftmap.bu.edu/.
Figures


References
-
- Allen K, et al. An experimental approach to mapping the binding surfaces of crystalline proteins. J. Phys. Chem. 1996;100:2605–2611.
-
- An JH, et al. Pocketome via comprehensive identification and classification of ligand binding envelopes. Mol. Cell. Proteomics. 2005;4:752–761. - PubMed
-
- Brooks BR, et al. CHARMM: a program for macromolecular energy, minimization, and dynamics calculations. J. Comp. Chem. 1983;4:187–217.
-
- Caflisch A, et al. Multiple copy simultaneous search and construction of ligands in binding sites: application to inhibitors of HIV-1 aspartic proteinase. J. Med. Chem. 1993;36:2142–2167. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

