A catalytic metal ion interacts with the cleavage Site G.U wobble in the HDV ribozyme
- PMID: 19178151
- PMCID: PMC2645270
- DOI: 10.1021/bi8020108
A catalytic metal ion interacts with the cleavage Site G.U wobble in the HDV ribozyme
Abstract
The HDV ribozyme self-cleaves by a chemical mechanism involving general acid-base catalysis to generate 2',3'-cyclic phosphate and 5'-hydroxyl termini. Biochemical studies from several laboratories have implicated C75 as the general acid and hydrated magnesium as the general base. We have previously shown that C75 has a pK(a) shifted >2 pH units toward neutrality [Gong, B., Chen, J. H., Chase, E., Chadalavada, D. M., Yajima, R., Golden, B. L., Bevilacqua, P. C., and Carey, P. R. (2007) J. Am. Chem. Soc. 129, 13335-13342], while in crystal structures, it is well-positioned for proton transfer. However, no evidence for a hydrated magnesium poised to serve as a general base in the reaction has been observed in high-resolution crystal structures of various reaction states and mutants. Herein, we use solution kinetic experiments and parallel Raman crystallographic studies to examine the effects of pH on the rate and Mg(2+) binding properties of wild-type and 7-deazaguanosine mutants of the HDV ribozyme. These data suggest that a previously unobserved hydrated magnesium ion interacts with N7 of the cleavage site G.U wobble base pair. Integrating this metal ion binding site with the available crystal structures provides a new three-dimensional model for the active site of the ribozyme that accommodates all available biochemical data and appears competent for catalysis. The position of this metal is consistent with a role of a magnesium-bound hydroxide as a general base as dictated by biochemical data.
Figures
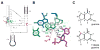
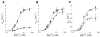
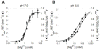

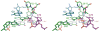
References
-
- Lai MM. The molecular biology of hepatitis delta virus. Annu Rev Biochem. 1995;64:259–286. - PubMed
-
- Been MD, Wickham GS. Self-cleaving ribozymes of hepatitis delta virus RNA. Eur J Biochem. 1997;247:741–753. - PubMed
-
- Salehi-Ashtiani K, Luptak A, Litovchick A, Szostak JW. A genomewide search for ribozymes reveals an HDV-like sequence in the human CPEB3 gene. Science. 2006;313:1788–1792. - PubMed
-
- Ferre-D’Amare AR, Zhou K, Doudna JA. Crystal structure of a hepatitis delta virus ribozyme. Nature. 1998;395:567–574. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

