Genomic islands: tools of bacterial horizontal gene transfer and evolution
- PMID: 19178566
- PMCID: PMC2704930
- DOI: 10.1111/j.1574-6976.2008.00136.x
Genomic islands: tools of bacterial horizontal gene transfer and evolution
Abstract
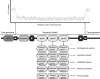
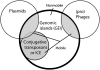
Bacterial genomes evolve through mutations, rearrangements or horizontal gene transfer. Besides the core genes encoding essential metabolic functions, bacterial genomes also harbour a number of accessory genes acquired by horizontal gene transfer that might be beneficial under certain environmental conditions. The horizontal gene transfer contributes to the diversification and adaptation of microorganisms, thus having an impact on the genome plasticity. A significant part of the horizontal gene transfer is or has been facilitated by genomic islands (GEIs). GEIs are discrete DNA segments, some of which are mobile and others which are not, or are no longer mobile, which differ among closely related strains. A number of GEIs are capable of integration into the chromosome of the host, excision, and transfer to a new host by transformation, conjugation or transduction. GEIs play a crucial role in the evolution of a broad spectrum of bacteria as they are involved in the dissemination of variable genes, including antibiotic resistance and virulence genes leading to generation of hospital 'superbugs', as well as catabolic genes leading to formation of new metabolic pathways. Depending on the composition of gene modules, the same type of GEIs can promote survival of pathogenic as well as environmental bacteria.
Figures


References
-
- Beaber JW, Hochhut B, Waldor MK. SOS response promotes horizontal dissemination of antibiotic resistance genes. Nature. 2004;427:72–74. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

