An Entry/Gateway cloning system for general expression of genes with molecular tags in Drosophila melanogaster
- PMID: 19178707
- PMCID: PMC2654426
- DOI: 10.1186/1471-2121-10-8
An Entry/Gateway cloning system for general expression of genes with molecular tags in Drosophila melanogaster
Abstract
Background: Tagged fusion proteins are priceless tools for monitoring the activities of biomolecules in living cells. However, over-expression of fusion proteins sometimes leads to the unwanted lethality or developmental defects. Therefore, vectors that can express tagged proteins at physiological levels are desirable tools for studying dosage-sensitive proteins. We developed a set of Entry/Gateway vectors for expressing fluorescent fusion proteins in Drosophila melanogaster. The vectors were used to generate fluorescent CP190 which is a component of the gypsy chromatin insulator. We used the fluorescent CP190 to study the dynamic movement of related chromatin insulators in living cells.
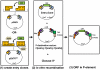
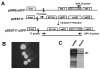
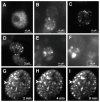
Results: The Entry/Gateway system is a timesaving technique for quickly generating expression constructs of tagged fusion proteins. We described in this study an Entry/Gateway based system, which includes six P-element destination vectors (P-DEST) for expressing tagged proteins (eGFP, mRFP, or myc) in Drosophila melanogaster and a TA-based cloning vector for generating entry clones from unstable DNA sequences. We used the P-DEST vectors to express fluorecent CP190 at tolerable levels. Expression of CP190 using the UAS/Gal4 system, instead, led to either lethality or underdeveloped tissues. The expressed eGFP- or mRFP-tagged CP190 proteins are fully functional and rescued the lethality of the homozygous CP190 mutation. We visualized a wide range of CP190 distribution patterns in living cell nuclei, from thousands of tiny particles to less than ten giant ones, which likely reflects diverse organization of higher-order chromatin structures. We also visualized the fusion of multiple smaller insulator bodies into larger aggregates in living cells, which is likely reflective of the dynamic activities of reorganization of chromatin in living nuclei.
Conclusion: We have developed an efficient cloning system for expressing dosage-sensitive proteins in Drosophila melanogaster. This system successfully expresses functional fluorescent CP190 fusion proteins. The fluorescent CP190 proteins exist in insulator bodies of various numbers and sizes among cells from multiple living tissues. Furthermore, live imaging of the movements of these fluorescent-tagged proteins suggests that the assembly and disassembly of insulator bodies are normal activities in living cells and may be directed for regulating transcription.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

